CXM: a new tool for mapping breast cancer risk in the tumor microenvironment
- PMID: 25172839
- PMCID: PMC4247541
- DOI: 10.1158/0008-5472.CAN-13-3212
CXM: a new tool for mapping breast cancer risk in the tumor microenvironment
Abstract
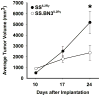
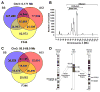
The majority of causative variants in familial breast cancer remain unknown. Of the known risk variants, most are tumor cell autonomous, and little attention has been paid yet to germline variants that may affect the tumor microenvironment. In this study, we developed a system called the Consomic Xenograft Model (CXM) to map germline variants that affect only the tumor microenvironment. In CXM, human breast cancer cells are orthotopically implanted into immunodeficient consomic strains and tumor metrics are quantified (e.g., growth, vasculogenesis, and metastasis). Because the strain backgrounds vary, whereas the malignant tumor cells do not, any observed changes in tumor progression are due to genetic differences in the nonmalignant microenvironment. Using CXM, we defined genetic variants on rat chromosome 3 that reduced relative tumor growth and hematogenous metastasis in the SS.BN3(IL2Rγ) consomic model compared with the SS(IL2Rγ) parental strain. Paradoxically, these effects occurred despite an increase in the density of tumor-associated blood vessels. In contrast, lymphatic vasculature and lymphogenous metastasis were unaffected by the SS.BN3(IL2Rγ) background. Through comparative mapping and whole-genome sequence analysis, we narrowed candidate variants on rat chromosome 3 to six genes with a priority for future analysis. Collectively, our results establish the utility of CXM to localize genetic variants affecting the tumor microenvironment that underlie differences in breast cancer risk.
©2014 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nat Genet. 2000;26:411–414. - PubMed
-
- Pharoah PD, Antoniou A, Bobrow M, Zimmern RL, Easton DF, Ponder BA. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

