Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa
- PMID: 25172852
- PMCID: PMC4249032
- DOI: 10.1128/AEM.01237-14
Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa
Abstract
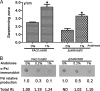
Biofilm formation is a complex process in which many factors are involved. Bacterial swarming motility and exopolysaccharides both contribute to biofilm formation, yet it is unclear how bacteria coordinate swarming motility and exopolysaccharide production. Psl and Pel are two key biofilm matrix exopolysaccharides in Pseudomonas aeruginosa. This opportunistic pathogen has three types of motility, swimming, twitching, and swarming. In this study, we found that elevated Psl and/or Pel production reduced the swarming motility of P. aeruginosa but had little effect on swimming and twitching. The reduction was due to decreased rhamnolipid production with no relation to the transcription of rhlAB, two key genes involved in the biosynthesis of rhamnolipids. Rhamnolipid-negative rhlR and rhlAB mutants synthesized more Psl, whereas exopolysaccharide-deficient strains exhibited a hyperswarming phenotype. These results suggest that competition for common sugar precursors catalyzed by AlgC could be a tactic for P. aeruginosa to balance the synthesis of exopolysaccharides and rhamnolipids and to control bacterial motility and biofilm formation inversely because the biosynthesis of rhamnolipids, Psl, and Pel requires AlgC to provide the sugar precursors and an additional algC gene enhances the biosynthesis of Psl and rhamnolipids. In addition, our data indicate that the increase in RhlI/RhlR expression attenuated Psl production. This implied that the quorum-sensing signals could regulate exopolysaccharide biosynthesis indirectly in bacterial communities. In summary, this study represents a mechanism that bacteria utilize to coordinate swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production, which is critical for biofilm formation and bacterial survival in the environment.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
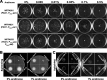
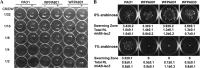
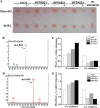
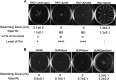
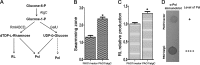
Similar articles
-
A Survival Strategy for Pseudomonas aeruginosa That Uses Exopolysaccharides To Sequester and Store Iron To Stimulate Psl-Dependent Biofilm Formation.Appl Environ Microbiol. 2016 Oct 14;82(21):6403-6413. doi: 10.1128/AEM.01307-16. Print 2016 Nov 1. Appl Environ Microbiol. 2016. PMID: 27565622 Free PMC article.
-
Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated.Environ Microbiol. 2012 Aug;14(8):1995-2005. doi: 10.1111/j.1462-2920.2012.02753.x. Epub 2012 Apr 19. Environ Microbiol. 2012. PMID: 22513190 Free PMC article.
-
Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885).PLoS Pathog. 2009 Jun;5(6):e1000483. doi: 10.1371/journal.ppat.1000483. Epub 2009 Jun 19. PLoS Pathog. 2009. PMID: 19543378 Free PMC article.
-
Regulation of Biofilm Exopolysaccharide Biosynthesis and Degradation in Pseudomonas aeruginosa.Annu Rev Microbiol. 2022 Sep 8;76:413-433. doi: 10.1146/annurev-micro-041320-111355. Epub 2022 Jun 2. Annu Rev Microbiol. 2022. PMID: 35655342 Review.
-
Role of polysaccharides in Pseudomonas aeruginosa biofilm development.Curr Opin Microbiol. 2007 Dec;10(6):644-8. doi: 10.1016/j.mib.2007.09.010. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981495 Free PMC article. Review.
Cited by
-
Beyond the Wall: Exopolysaccharides in the Biofilm Lifestyle of Pathogenic and Beneficial Plant-Associated Pseudomonas.Microorganisms. 2021 Feb 21;9(2):445. doi: 10.3390/microorganisms9020445. Microorganisms. 2021. PMID: 33670010 Free PMC article. Review.
-
EGCG-Mediated Potential Inhibition of Biofilm Development and Quorum Sensing in Pseudomonas aeruginosa.Int J Mol Sci. 2021 May 6;22(9):4946. doi: 10.3390/ijms22094946. Int J Mol Sci. 2021. PMID: 34066609 Free PMC article.
-
Sodium Selenite Enhances Antibiotics Sensitivity of Pseudomonas aeruginosa and Deceases Its Pathogenicity by Inducing Oxidative Stress and Inhibiting Quorum Sensing System.Antioxidants (Basel). 2021 Nov 24;10(12):1873. doi: 10.3390/antiox10121873. Antioxidants (Basel). 2021. PMID: 34942975 Free PMC article.
-
Integrated Comparative Genomic Analysis and Phenotypic Profiling of Pseudomonas aeruginosa Isolates From Crude Oil.Front Microbiol. 2020 Mar 31;11:519. doi: 10.3389/fmicb.2020.00519. eCollection 2020. Front Microbiol. 2020. PMID: 32300337 Free PMC article.
-
Genomic characterization of a polyvalent hydrocarbonoclastic bacterium Pseudomonas sp. strain BUN14.Sci Rep. 2021 Apr 14;11(1):8124. doi: 10.1038/s41598-021-87487-2. Sci Rep. 2021. PMID: 33854112 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

