Mutation of the Erwinia amylovora argD gene causes arginine auxotrophy, nonpathogenicity in apples, and reduced virulence in pears
- PMID: 25172854
- PMCID: PMC4249043
- DOI: 10.1128/AEM.02404-14
Mutation of the Erwinia amylovora argD gene causes arginine auxotrophy, nonpathogenicity in apples, and reduced virulence in pears
Abstract
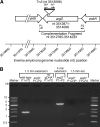
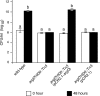
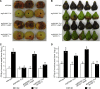
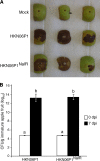
Fire blight is caused by Erwinia amylovora and is the most destructive bacterial disease of apples and pears worldwide. In this study, we found that E. amylovora argD(1000)::Tn5, an argD Tn5 transposon mutant that has the Tn5 transposon inserted after nucleotide 999 in the argD gene-coding region, was an arginine auxotroph that did not cause fire blight in apple and had reduced virulence in immature pear fruits. The E. amylovora argD gene encodes a predicted N-acetylornithine aminotransferase enzyme, which is involved in the production of the amino acid arginine. A plasmid-borne copy of the wild-type argD gene complemented both the nonpathogenic and the arginine auxotrophic phenotypes of the argD(1000)::Tn5 mutant. However, even when mixed with virulent E. amylovora cells and inoculated onto immature apple fruit, the argD(1000)::Tn5 mutant still failed to grow, while the virulent strain grew and caused disease. Furthermore, the pCR2.1-argD complementation plasmid was stably maintained in the argD(1000)::Tn5 mutant growing in host tissues without any antibiotic selection. Therefore, the pCR2.1-argD complementation plasmid could be useful for the expression of genes, markers, and reporters in E. amylovora growing in planta, without concern about losing the plasmid over time. The ArgD protein cannot be considered an E. amylovora virulence factor because the argD(1000)::Tn5 mutant was auxotrophic and had a primary metabolism defect. Nevertheless, these results are informative about the parasitic nature of the fire blight disease interaction, since they indicate that E. amylovora cannot obtain sufficient arginine from apple and pear fruit tissues or from apple vegetative tissues, either at the beginning of the infection process or after the infection has progressed to an advanced state.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Erwinia amylovora pyrC mutant causes fire blight despite pyrimidine auxotrophy.Lett Appl Microbiol. 2015 Jun;60(6):572-9. doi: 10.1111/lam.12417. Epub 2015 Apr 16. Lett Appl Microbiol. 2015. PMID: 25789570
-
Erwinia amylovora Auxotrophic Mutant Exometabolomics and Virulence on Apples.Appl Environ Microbiol. 2019 Jul 18;85(15):e00935-19. doi: 10.1128/AEM.00935-19. Print 2019 Aug 1. Appl Environ Microbiol. 2019. PMID: 31152019 Free PMC article.
-
Virulence Genetics of an Erwinia amylovora Putative Polysaccharide Transporter Family Member.J Bacteriol. 2020 Oct 22;202(22):e00390-20. doi: 10.1128/JB.00390-20. Print 2020 Oct 22. J Bacteriol. 2020. PMID: 32839177 Free PMC article.
-
Current Situation of Fire Blight in China.Phytopathology. 2023 Dec;113(12):2143-2151. doi: 10.1094/PHYTO-05-23-0170-RVW. Epub 2023 Dec 26. Phytopathology. 2023. PMID: 37505073 Review.
-
Molecular genetics of Erwinia amylovora involved in the development of fire blight.FEMS Microbiol Lett. 2005 Dec 15;253(2):185-92. doi: 10.1016/j.femsle.2005.09.051. Epub 2005 Oct 13. FEMS Microbiol Lett. 2005. PMID: 16253442 Review.
Cited by
-
Disruption of the carA gene in Pseudomonas syringae results in reduced fitness and alters motility.BMC Microbiol. 2016 Aug 24;16(1):194. doi: 10.1186/s12866-016-0819-z. BMC Microbiol. 2016. PMID: 27558694 Free PMC article.
-
Comparative transcriptome analysis of a lowly virulent strain of Erwinia amylovora in shoots of two apple cultivars - susceptible and resistant to fire blight.BMC Genomics. 2017 Nov 13;18(1):868. doi: 10.1186/s12864-017-4251-z. BMC Genomics. 2017. PMID: 29132313 Free PMC article.
-
The stringent response regulator (p) ppGpp mediates virulence gene expression and survival in Erwinia amylovora.BMC Genomics. 2020 Mar 30;21(1):261. doi: 10.1186/s12864-020-6699-5. BMC Genomics. 2020. PMID: 32228459 Free PMC article.
-
Transcription Factor VdCf2 Regulates Growth, Pathogenicity, and the Expression of a Putative Secondary Metabolism Gene Cluster in Verticillium dahliae.Appl Environ Microbiol. 2022 Nov 22;88(22):e0138522. doi: 10.1128/aem.01385-22. Epub 2022 Nov 7. Appl Environ Microbiol. 2022. PMID: 36342142 Free PMC article.
-
Identification of novel virulence factors in Erwinia amylovora through temporal transcriptomic analysis of infected apple flowers under field conditions.Mol Plant Pathol. 2022 Jun;23(6):855-869. doi: 10.1111/mpp.13199. Epub 2022 Mar 4. Mol Plant Pathol. 2022. PMID: 35246928 Free PMC article.
References
-
- Burrill TJ. 1880. Anthrax of fruit trees; or the so-called fire blight of pear, and twig blight of apple trees. Am. Assoc. Adv. Sci. Proc. 29:583–597.
-
- Arthur JC. 1885. Proof that bacteria are the direct cause of the disease in trees known as pear blight. Am. Assoc. Adv. Sci. Proc. 34:294–298.
-
- Coxe W. 1817. A view of the cultivation of fruit trees, and the management of orchards and cider. M. Carey & Son, Philadelphia, PA.
-
- Momol MT, Yegen O. 1993. Fire blight in Turkey: 1985-1992. Acta Hort. 338:37–39.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

