Genetic differentiation of hypothalamus parentally biased transcripts in populations of the house mouse implicate the Prader-Willi syndrome imprinted region as a possible source of behavioral divergence
- PMID: 25172960
- PMCID: PMC4245819
- DOI: 10.1093/molbev/msu257
Genetic differentiation of hypothalamus parentally biased transcripts in populations of the house mouse implicate the Prader-Willi syndrome imprinted region as a possible source of behavioral divergence
Erratum in
-
Genetic Differentiation of Hypothalamus Parentally Biased Transcripts in Populations of the House Mouse Implicate the Prader-Willi Syndrome Imprinted Region as a Possible Source of Behavioral Divergence.Mol Biol Evol. 2015 Jul;32(7):1914-5. doi: 10.1093/molbev/msv108. Epub 2015 May 19. Mol Biol Evol. 2015. PMID: 25989982 Free PMC article. No abstract available.
Abstract
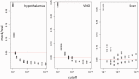
Parentally biased expression of transcripts (genomic imprinting) in adult tissues, including the brain, can influence and possibly drive the evolution of behavioral traits. We have previously found that paternally determined cues are involved in population-specific mate choice decisions between two populations of the Western house mouse (Mus musculus domesticus). Here, we ask whether this could be mediated by genomically imprinted transcripts that are subject to fast differentiation between these populations. We focus on three organs that are of special relevance for mate choice and behavior: The vomeronasal organ (VNO), the hypothalamus, and the liver. To first identify candidate transcripts at a genome-wide scale, we used reciprocal crosses between M. m. domesticus and M. m. musculus inbred strains and RNA sequencing of the respective tissues. Using a false discovery cutoff derived from mock reciprocal cross comparisons, we find a total of 66 imprinted transcripts, 13 of which have previously not been described as imprinted. The largest number of imprinted transcripts were found in the hypothalamus; fewer were found in the VNO, and the least were found in the liver. To assess molecular differentiation and imprinting in the wild-derived M. m. domesticus populations, we sequenced the RNA of the hypothalamus from individuals of these populations. This confirmed the presence of the above identified transcripts also in wild populations and allowed us to search for those that show a high genetic differentiation between these populations. Our results identify the Ube3a-Snrpn imprinted region on chromosome 7 as a region that encompasses the largest number of previously not described transcripts with paternal expression bias, several of which are at the same time highly differentiated. For four of these, we confirmed their imprinting status via single nucleotide polymorphism-specific pyrosequencing assays with RNA from reciprocal crosses. In addition, we find the paternally expressed Peg13 transcript within the Trappc9 gene region on chromosome 15 to be highly differentiated. Interestingly, both regions have been implicated in Prader-Willi nervous system disorder phenotypes in humans. We suggest that these genomically imprinted regions are candidates for influencing the population-specific mate-choice in mice.
Keywords: Mus domesticus; Mus musculus; RNASeq; genetic differentiation; hypothalamus; imprinting; liver; vomeronasal organ.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
Similar articles
-
Identification of novel imprinted transcripts in the Prader-Willi syndrome and Angelman syndrome deletion region: further evidence for regional imprinting control.Am J Hum Genet. 2000 Mar;66(3):848-58. doi: 10.1086/302817. Am J Hum Genet. 2000. PMID: 10712201 Free PMC article.
-
An unexpected function of the Prader-Willi syndrome imprinting center in maternal imprinting in mice.PLoS One. 2012;7(4):e34348. doi: 10.1371/journal.pone.0034348. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496793 Free PMC article.
-
Imprinted genes in the Prader-Willi deletion.Novartis Found Symp. 1998;214:264-75; discussion 275-9. doi: 10.1002/9780470515501.ch16. Novartis Found Symp. 1998. PMID: 9601023 Review.
-
The Prader-Willi syndrome murine imprinting center is not involved in the spatio-temporal transcriptional regulation of the Necdin gene.BMC Genet. 2005 Jan 5;6:1. doi: 10.1186/1471-2156-6-1. BMC Genet. 2005. PMID: 15634360 Free PMC article.
-
Genetic abnormalities in Prader-Willi syndrome and lessons from mouse models.Acta Paediatr Suppl. 1999 Dec;88(433):99-104. doi: 10.1111/j.1651-2227.1999.tb14414.x. Acta Paediatr Suppl. 1999. PMID: 10626556 Review.
Cited by
-
ISoLDE: a data-driven statistical method for the inference of allelic imbalance in datasets with reciprocal crosses.Bioinformatics. 2020 Jan 15;36(2):504-513. doi: 10.1093/bioinformatics/btz564. Bioinformatics. 2020. PMID: 31350542 Free PMC article.
-
Quantifying Genomic Imprinting at Tissue and Cell Resolution in the Brain.Epigenomes. 2020 Sep 4;4(3):21. doi: 10.3390/epigenomes4030021. Epigenomes. 2020. PMID: 34968292 Free PMC article. Review.
-
Comparison of gut microflora of donkeys in high and low altitude areas.Front Microbiol. 2022 Sep 26;13:964799. doi: 10.3389/fmicb.2022.964799. eCollection 2022. Front Microbiol. 2022. PMID: 36225357 Free PMC article.
-
Replicate sequencing libraries are important for quantification of allelic imbalance.Nat Commun. 2021 Jun 7;12(1):3370. doi: 10.1038/s41467-021-23544-8. Nat Commun. 2021. PMID: 34099647 Free PMC article.
-
The imprinted lncRNA Peg13 regulates sexual preference and the sex-specific brain transcriptome in mice.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2022172118. doi: 10.1073/pnas.2022172118. Proc Natl Acad Sci U S A. 2021. Retraction in: Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2300529120. doi: 10.1073/pnas.2300529120. PMID: 33658376 Free PMC article. Retracted.
References
-
- Arnaud P, Monk D, Hitchins M, Gordon E, Dean W, Beechey CV, Peters J, Craigen W, Preece M, Stanier P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12:1005–1019. - PubMed
-
- Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader–Willi syndrome. Genet Med. 2012;14:10–26. - PubMed
-
- Chai JH, Locke DP, Ohta T, Greally JM, Nicholls RD. Retrotransposed genes such as Frat3 in the mouse Chromosome 7C Prader-Willi syndrome region acquire the imprinted status of their insertion site. Mamm Genome. 2001;12:813–821. - PubMed
-
- Court F, Camprubi C, Garcia CV, Guillaumet-Adkins A, Sparago A, Seruggia D, Sandoval J, Esteller M, Martin-Trujillo A, Riccio A, et al. The PEG13-DMR and brain-specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus. Epigenetics Chromatin. 2014;7:5. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

