Drosophila melanogaster cellular repressor of E1A-stimulated genes is a lysosomal protein essential for fly development
- PMID: 25173815
- PMCID: PMC4331662
- DOI: 10.1016/j.bbamcr.2014.08.012
Drosophila melanogaster cellular repressor of E1A-stimulated genes is a lysosomal protein essential for fly development
Abstract
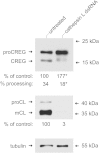
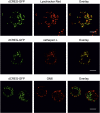
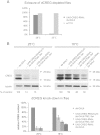
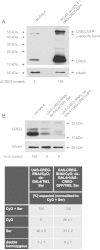
Mammalian cellular repressor of E1A-stimulated genes is a lysosomal glycoprotein implicated in cellular growth and differentiation. The genome of the fruit fly Drosophila melanogaster encodes a putative orthologue (dCREG), suggesting evolutionarily conserved physiological functions of this protein. In D. melanogaster S2 cells, dCREG was found to localize in lysosomes. Further studies revealed that intracellular dCREG is subject of proteolytic maturation. Processing and turnover could be substantially reduced by RNAi-mediated silencing of cathepsin L. In contrast to mammalian cells, lysosomal delivery of dCREG does not depend on its carbohydrate moiety. Furthermore, depletion of the putative D. melanogaster lysosomal sorting receptor lysosomal enzyme receptor protein did not compromise cellular retention of dCREG. We also investigated the developmental consequences of dCREG ablation in whole D. melanogaster flies. Ubiquitous depletion of dCREG proved lethal at the late pupal stage once a knock-down efficiency of >95% was achieved. These results demonstrate that dCREG is essential for proper completion of fly development.
Keywords: CREG; Cathepsin; Lysosome; Mannose 6-phosphate; Protein targeting; Proteolytic maturation.
Copyright © 2014. Published by Elsevier B.V.
Figures





References
-
- Veal E., Groisman R., Eisenstein M., Gill G. The secreted glycoprotein CREG enhances differentiation of NTERA-2 human embryonal carcinoma cells. Oncogene. 2000;19:2120–2128. - PubMed
-
- Han Y.L., Guo P., Sun M.Y., Guo L., Luan B., Kang J., Yan C.H., Li S.H. Secreted CREG inhibits cell proliferation mediated by mannose 6-phosphate/insulin-like growth factor II receptor in NIH3T3 fibroblasts. Genes Cells. 2008;13:977–986. - PubMed
-
- Xu L., Liu J.M., Chen L.Y. CREG, a new regulator of ERK1/2 in cardiac hypertrophy. J. Hypertens. 2004;22:1579–1587. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

