Exploiting natural variation of secondary metabolism identifies a gene controlling the glycosylation diversity of dihydroxybenzoic acids in Arabidopsis thaliana
- PMID: 25173843
- PMCID: PMC4224165
- DOI: 10.1534/genetics.114.168690
Exploiting natural variation of secondary metabolism identifies a gene controlling the glycosylation diversity of dihydroxybenzoic acids in Arabidopsis thaliana
Abstract
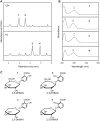
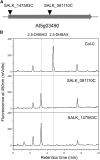
Plant secondary metabolism is an active research area because of the unique and important roles the specialized metabolites have in the interaction of plants with their biotic and abiotic environment, the diversity and complexity of the compounds and their importance to human medicine. Thousands of natural accessions of Arabidopsis thaliana characterized with increasing genomic precision are available, providing new opportunities to explore the biochemical and genetic mechanisms affecting variation in secondary metabolism within this model species. In this study, we focused on four aromatic metabolites that were differentially accumulated among 96 Arabidopsis natural accessions as revealed by leaf metabolic profiling. Using UV, mass spectrometry, and NMR data, we identified these four compounds as different dihydroxybenzoic acid (DHBA) glycosides, namely 2,5-dihydroxybenzoic acid (gentisic acid) 5-O-β-D-glucoside, 2,3-dihydroxybenzoic acid 3-O-β-D-glucoside, 2,5-dihydroxybenzoic acid 5-O-β-D-xyloside, and 2,3-dihydroxybenzoic acid 3-O-β-D-xyloside. Quantitative trait locus (QTL) mapping using recombinant inbred lines generated from C24 and Col-0 revealed a major-effect QTL controlling the relative proportion of xylosides vs. glucosides. Association mapping identified markers linked to a gene encoding a UDP glycosyltransferase gene. Analysis of Transfer DNA (T-DNA) knockout lines verified that this gene is required for DHBA xylosylation in planta and recombinant protein was able to xylosylate DHBA in vitro. This study demonstrates that exploiting natural variation of secondary metabolism is a powerful approach for gene function discovery.
Keywords: Arabidopsis; UDP glycosyltransferase (UGT); dihydroxybenzoic acid glycosides; natural variation; secondary metabolism.
Copyright © 2014 by the Genetics Society of America.
Figures






Similar articles
-
Identification of a residue responsible for UDP-sugar donor selectivity of a dihydroxybenzoic acid glycosyltransferase from Arabidopsis natural accessions.Plant J. 2017 Jan;89(2):195-203. doi: 10.1111/tpj.13271. Epub 2016 Sep 15. Plant J. 2017. PMID: 27411741
-
Modulation of Plant Salicylic Acid-Associated Immune Responses via Glycosylation of Dihydroxybenzoic Acids.Plant Physiol. 2018 Apr;176(4):3103-3119. doi: 10.1104/pp.17.01530. Epub 2018 Feb 26. Plant Physiol. 2018. PMID: 29483147 Free PMC article.
-
Accumulation of isochorismate-derived 2,3-dihydroxybenzoic 3-O-beta-D-xyloside in arabidopsis resistance to pathogens and ageing of leaves.J Biol Chem. 2010 Aug 13;285(33):25654-65. doi: 10.1074/jbc.M109.092569. Epub 2010 Jun 10. J Biol Chem. 2010. PMID: 20538606 Free PMC article.
-
Naturally occurring genetic variation in Arabidopsis thaliana.Annu Rev Plant Biol. 2004;55:141-72. doi: 10.1146/annurev.arplant.55.031903.141605. Annu Rev Plant Biol. 2004. PMID: 15377217 Review.
-
Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology.Ann Bot. 2007 Jun;99(6):1043-54. doi: 10.1093/aob/mcl281. Epub 2007 Jan 26. Ann Bot. 2007. PMID: 17259228 Free PMC article. Review.
Cited by
-
Salicylic Acid Is Required for Broad-Spectrum Disease Resistance in Rice.Int J Mol Sci. 2022 Jan 25;23(3):1354. doi: 10.3390/ijms23031354. Int J Mol Sci. 2022. PMID: 35163275 Free PMC article.
-
Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data.Sci Rep. 2016 Jul 1;6:29033. doi: 10.1038/srep29033. Sci Rep. 2016. PMID: 27363486 Free PMC article.
-
S5H/DMR6 Encodes a Salicylic Acid 5-Hydroxylase That Fine-Tunes Salicylic Acid Homeostasis.Plant Physiol. 2017 Nov;175(3):1082-1093. doi: 10.1104/pp.17.00695. Epub 2017 Sep 12. Plant Physiol. 2017. PMID: 28899963 Free PMC article.
-
Discovery of a novel amino acid racemase through exploration of natural variation in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11726-31. doi: 10.1073/pnas.1503272112. Epub 2015 Aug 31. Proc Natl Acad Sci U S A. 2015. PMID: 26324904 Free PMC article.
-
QT-GWAS: A novel method for unveiling biosynthetic loci affecting qualitative metabolic traits.Mol Plant. 2023 Jul 3;16(7):1212-1227. doi: 10.1016/j.molp.2023.06.004. Epub 2023 Jun 21. Mol Plant. 2023. PMID: 37349988 Free PMC article.
References
-
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H. M., et al. , 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Balandrin M. F., Klocke J. A., Wurtele E. S., Bollinger W. H., 1985. Natural plant chemicals: sources of industrial and medicinal materials. Science 228: 1154–1160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

