Mammary stem cells have myoepithelial cell properties
- PMID: 25173976
- PMCID: PMC4183554
- DOI: 10.1038/ncb3025
Mammary stem cells have myoepithelial cell properties
Abstract
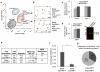
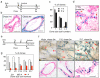
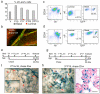
Contractile myoepithelial cells dominate the basal layer of the mammary epithelium and are considered to be differentiated cells. However, we observe that up to 54% of single basal cells can form colonies when seeded into adherent culture in the presence of agents that disrupt actin-myosin interactions, and on average, 65% of the single-cell-derived basal colonies can repopulate a mammary gland when transplanted in vivo. This indicates that a high proportion of basal myoepithelial cells can give rise to a mammary repopulating unit (MRU). We demonstrate that myoepithelial cells, flow-sorted using two independent myoepithelial-specific reporter strategies, have MRU capacity. Using an inducible lineage-tracing approach we follow the progeny of myoepithelial cells that express α-smooth muscle actin and show that they function as long-lived lineage-restricted stem cells in the virgin state and during pregnancy.
Figures



References
-
- Chepko G, et al. Differential alteration of stem and other cell populations in ducts and lobules of TGF[alpha] and c-Myc transgenic mouse mammary epithelium. Tissue and Cell. 2005;37:393–412. - PubMed
-
- Smith GH, Medina D. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. Journal of Cell Science. 1988;90:173–183. - PubMed
-
- Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

