A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis
- PMID: 25175028
- PMCID: PMC4169771
- DOI: 10.1016/j.molcel.2014.07.017
A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis
Abstract
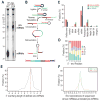
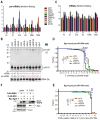
MicroRNAs (miRNAs) are essential for regulation of gene expression. Though numerous miRNAs have been identified by high-throughput sequencing, few precursor miRNAs (pre-miRNAs) are experimentally validated. Here we report a strategy for constructing high-throughput sequencing libraries enriched for full-length pre-miRNAs. We find widespread and extensive uridylation of Argonaute (Ago)-bound pre-miRNAs, which is primarily catalyzed by two terminal uridylyltransferases: TUT7 and TUT4. Uridylation by TUT7/4 not only polishes pre-miRNA 3' ends, but also facilitates their degradation by the exosome, preventing clogging of Ago with defective species. We show that the exosome exploits distinct substrate preferences of DIS3 and RRP6, its two catalytic subunits, to distinguish productive from defective pre-miRNAs. Furthermore, we identify a positive feedback loop formed by the exosome and TUT7/4 in triggering uridylation and degradation of Ago-bound pre-miRNAs. Our study reveals a pre-miRNA surveillance system that comprises TUT7, TUT4, and the exosome in quality control of miRNA synthesis.
Copyright © 2014 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





References
-
- Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

