High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules
- PMID: 25176140
- PMCID: PMC4241113
- DOI: 10.1002/pro.2543
High-resolution crystal structures of the photoreceptor glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with three and four-bound NAD molecules
Abstract
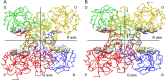
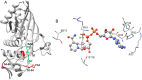
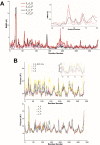
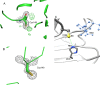
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the oxidative phosphorylation of d-glyceraldehyde 3-phosphate (G3P) into 1,3-diphosphoglycerate (BGP) in the presence of the NAD cofactor. GAPDH is an important drug target because of its central role in glycolysis, and nonglycolytic processes such as nuclear RNA transport, DNA replication/repair, membrane fusion and cellular apoptosis. Recent studies found that GAPDH participates in the development of diabetic retinopathy and its progression after the cessation of hyperglycemia. Here, we report two structures for native bovine photoreceptor GAPDH as a homotetramer with differing occupancy by NAD, bGAPDH(NAD)4 , and bGAPDH(NAD)3 . The bGAPDH(NAD)4 was solved at 1.52 Å, the highest resolution for GAPDH. Structural comparison of the bGAPDH(NAD)4 and bGAPDH(NAD)3 models revealed novel details of conformational changes induced by cofactor binding, including a loop region (residues 54-56). Structure analysis of bGAPDH confirmed the importance of Phe34 in NAD binding, and demonstrated that Phe34 was stabilized in the presence of NAD but displayed greater mobility in its absence. The oxidative state of the active site Cys149 residue is regulated by NAD binding, because this residue was found oxidized in the absence of dinucleotide. The distance between Cys149 and His176 decreased upon NAD binding and Cys149 remained in a reduced state when NAD was bound. These findings provide an important structural step for understanding the mechanism of GAPDH activity in vision and its pathological role in retinopathies.
Keywords: NAD; S-nitrosylation; crystal structure; diabetic retinopathy; glyceraldehyde 3-phosphate dehydrogenase; oxidization; photoreceptor; structural comparison.
© 2014 The Protein Society.
Figures


References
-
- Seidler NW. Basic biology of GAPDH. Adv Exp Med Biol. 2013;985:1–36. - PubMed
-
- Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. - PubMed
-
- Sirover MA. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta. 2011;1810:741–751. - PubMed
-
- Padgett CM, Whorton AR. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol. 1995;269:C739–C749. - PubMed
-
- Mohr S, Stamler JS, Brune B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271:4209–4214. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

