Mapping the inner workings of the microbiome: genomic- and metagenomic-based study of metabolism and metabolic interactions in the human microbiome
- PMID: 25176148
- PMCID: PMC4252837
- DOI: 10.1016/j.cmet.2014.07.021
Mapping the inner workings of the microbiome: genomic- and metagenomic-based study of metabolism and metabolic interactions in the human microbiome
Abstract
The human gut microbiome is a major contributor to human metabolism and health, yet the metabolic processes that are carried out by various community members, the way these members interact with each other and with the host, and the impact of such interactions on the overall metabolic machinery of the microbiome have not yet been mapped. Here, we discuss recent efforts to study the metabolic inner workings of this complex ecosystem. We will specifically highlight two interrelated lines of work, the first aiming to deconvolve the microbiome and to characterize the metabolic capacity of various microbiome species and the second aiming to utilize computational modeling to infer and study metabolic interactions between these species.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
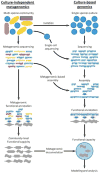

Similar articles
-
Determining microbial products and identifying molecular targets in the human microbiome.Cell Metab. 2014 Nov 4;20(5):731-741. doi: 10.1016/j.cmet.2014.10.003. Epub 2014 Nov 4. Cell Metab. 2014. PMID: 25440055 Free PMC article. Review.
-
Application of metagenomics in the human gut microbiome.World J Gastroenterol. 2015 Jan 21;21(3):803-14. doi: 10.3748/wjg.v21.i3.803. World J Gastroenterol. 2015. PMID: 25624713 Free PMC article. Review.
-
Gut metabolome meets microbiome: A methodological perspective to understand the relationship between host and microbe.Methods. 2018 Oct 1;149:3-12. doi: 10.1016/j.ymeth.2018.04.029. Epub 2018 Apr 30. Methods. 2018. PMID: 29715508 Review.
-
Modeling metabolism of the human gut microbiome.Curr Opin Biotechnol. 2018 Jun;51:90-96. doi: 10.1016/j.copbio.2017.12.005. Epub 2017 Dec 16. Curr Opin Biotechnol. 2018. PMID: 29258014 Review.
-
Metagenome-scale community metabolic modelling for understanding the role of gut microbiota in human health.Comput Biol Med. 2022 Oct;149:105997. doi: 10.1016/j.compbiomed.2022.105997. Epub 2022 Aug 19. Comput Biol Med. 2022. PMID: 36055158 Review.
Cited by
-
Metabolic network modeling of microbial communities.Wiley Interdiscip Rev Syst Biol Med. 2015 Sep-Oct;7(5):317-34. doi: 10.1002/wsbm.1308. Epub 2015 Jun 24. Wiley Interdiscip Rev Syst Biol Med. 2015. PMID: 26109480 Free PMC article. Review.
-
A quasi-paired cohort strategy reveals the impaired detoxifying function of microbes in the gut of autistic children.Sci Adv. 2020 Oct 21;6(43):eaba3760. doi: 10.1126/sciadv.aba3760. Print 2020 Oct. Sci Adv. 2020. PMID: 33087359 Free PMC article.
-
From cultured to uncultured genome sequences: metagenomics and modeling microbial ecosystems.Cell Mol Life Sci. 2015 Nov;72(22):4287-308. doi: 10.1007/s00018-015-2004-1. Epub 2015 Aug 9. Cell Mol Life Sci. 2015. PMID: 26254872 Free PMC article. Review.
-
Defining and Evaluating Microbial Contributions to Metabolite Variation in Microbiome-Metabolome Association Studies.mSystems. 2019 Dec 17;4(6):e00579-19. doi: 10.1128/mSystems.00579-19. mSystems. 2019. PMID: 31848305 Free PMC article.
-
Finding and identifying the viral needle in the metagenomic haystack: trends and challenges.Front Microbiol. 2015 Jan 7;5:739. doi: 10.3389/fmicb.2014.00739. eCollection 2014. Front Microbiol. 2015. PMID: 25610431 Free PMC article.
References
-
- Baker M. De novo genome assembly: what every biologist should know. Nature Methods. 2012;9:333–337.
-
- Baquero F, Nombela C. The microbiome as a human organ. Clinical Microbiology and Infection. 2012;18:2–4. - PubMed
-
- Borenstein E. Computational systems biology and in silico modeling of the human microbiome. Briefings in Bioinformatics. 2012;13:769–780. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

