Failsafe mechanisms couple division and DNA replication in bacteria
- PMID: 25176632
- PMCID: PMC4175050
- DOI: 10.1016/j.cub.2014.07.055
Failsafe mechanisms couple division and DNA replication in bacteria
Abstract
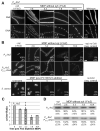
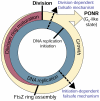
The past 20 years have seen tremendous advances in our understanding of the mechanisms underlying bacterial cytokinesis, particularly the composition of the division machinery and the factors controlling its assembly [1]. At the same time, we understand very little about the relationship between cell division and other cell-cycle events in bacteria. Here we report that inhibiting division in Bacillus subtilis and Staphylococcus aureus quickly leads to an arrest in the initiation of new rounds of DNA replication, followed by a complete arrest in cell growth. Arrested cells are metabolically active but are unable to initiate new rounds of either DNA replication or division when shifted to permissive conditions. Inhibiting DNA replication results in entry into a similar quiescent state in which cells are unable to resume growth or division when returned to permissive conditions. Our data suggest the presence of two failsafe mechanisms: one linking division to the initiation of DNA replication and another linking the initiation of DNA replication to division. These findings contradict the prevailing view of the bacterial cell cycle as a series of coordinated but uncoupled events. Importantly, the terminal nature of the cell-cycle arrest validates the bacterial cell-cycle machinery as an effective target for antimicrobial development.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


Comment in
-
Cell cycle: once out, never in again.Curr Biol. 2014 Sep 22;24(18):R841-R843. doi: 10.1016/j.cub.2014.07.067. Curr Biol. 2014. PMID: 25247357
References
-
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. - PubMed
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

