Submicroscopic deletion of 5q involving tumor suppressor genes (CTNNA1, HSPA9) and copy neutral loss of heterozygosity associated with TET2 and EZH2 mutations in a case of MDS with normal chromosome and FISH results
- PMID: 25177364
- PMCID: PMC4149311
- DOI: 10.1186/1755-8166-7-35
Submicroscopic deletion of 5q involving tumor suppressor genes (CTNNA1, HSPA9) and copy neutral loss of heterozygosity associated with TET2 and EZH2 mutations in a case of MDS with normal chromosome and FISH results
Abstract
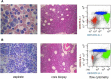
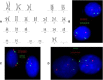
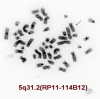
Advances in genome-wide molecular cytogenetics allow identification of novel submicroscopic DNA copy number alterations (aCNAs) and copy-neutral loss of heterozygosity (cnLOH) resulting in homozygosity for known gene mutations in myeloid neoplasms. We describe the use of an oligo-SNP array for genomic profiling of aCNA and cnLOH, together with sequence analysis of recurrently mutated genes, in a patient with myelodysplastic syndrome (MDS) presenting with normal karyotype and FISH results. Oligo-SNP array analysis revealed a hemizygous deletion of 896 kb at chromosome 5q31.2, representing the smallest 5q deletion reported to date. The deletion involved multiple genes, including two tumor suppressor candidate genes (CTNNA1 and HSPA9) that are associated with MDS/AML. The SNP-array study also detected 3 segments of somatic cnLOH: one involved the entire long arm of chromosome 4; the second involved the distal half of the long arm of chromosome 7, and the third encompassed the entire chromosome 22 (UPD 22). Sequence analysis revealed mutations in TET2 (4q), EZH2 (7q), ASXL1 (20q11.21), and RUNX1 (21q22.3). Coincidently, TET2 and EZH2 were located at segments of cnLOH resulting in their homozygosity. Loss of heterozygosity affecting these two chromosomes and mutations in TET2 and EZH2 are indicative of a myelodysplastic syndrome with a poor prognosis. Deletion of the tumor suppressor genes CTNNA1 and HSPA9 is also likely to contribute to a poor prognosis. Furthermore, the original cnLOHs in multiple chromosomes and additional cnLOH 14q in the follow-up study suggest genetic evolution of the disease and poor prognosis. This study attests to the fact that some patients with a myelodysplastic syndrome who exhibit a normal karyotype may have underlying genetic abnormalities detectable by chromosomal microarray and/or targeted mutation analyses.
Keywords: ASXL1; CTNNA1; Copy neutral loss of heterozygosity (cnLOH); EZH2; HSPA9; MDS; RUNX1; TET2; Uniparental disomy (UPD).
Figures



Similar articles
-
Integrated genomic profiling, therapy response, and survival in adult acute myelogenous leukemia.Clin Cancer Res. 2015 May 1;21(9):2045-56. doi: 10.1158/1078-0432.CCR-14-0921. Epub 2015 Feb 4. Clin Cancer Res. 2015. PMID: 25652455 Free PMC article.
-
Variant allele fraction or copy-neutral loss of heterozygosity? A comparison of testing platforms in the classification of myeloid neoplasia.J Hematop. 2025 Apr 23;18(1):21. doi: 10.1007/s12308-025-00636-8. J Hematop. 2025. PMID: 40266406
-
Copy number neutral loss of heterozygosity at 17p and homozygous mutations of TP53 are associated with complex chromosomal aberrations in patients newly diagnosed with myelodysplastic syndromes.Leuk Res. 2016 Mar;42:7-12. doi: 10.1016/j.leukres.2016.01.009. Epub 2016 Jan 24. Leuk Res. 2016. PMID: 26851439
-
The molecular pathogenesis of the myelodysplastic syndromes.Eur J Haematol. 2015 Jul;95(1):3-15. doi: 10.1111/ejh.12515. Epub 2015 Feb 20. Eur J Haematol. 2015. PMID: 25645650 Review.
-
Genetic abnormalities and pathophysiology of MDS.Int J Clin Oncol. 2019 Aug;24(8):885-892. doi: 10.1007/s10147-019-01462-6. Epub 2019 May 15. Int J Clin Oncol. 2019. PMID: 31093808 Review.
Cited by
-
The Role of CTNNA1 in Malignancies: An Updated Review.J Cancer. 2023 Jan 1;14(2):219-230. doi: 10.7150/jca.79236. eCollection 2023. J Cancer. 2023. PMID: 36741258 Free PMC article. Review.
-
Techniques for detecting chromosomal aberrations in myelodysplastic syndromes.Oncotarget. 2017 May 9;8(37):62716-62729. doi: 10.18632/oncotarget.17698. eCollection 2017 Sep 22. Oncotarget. 2017. PMID: 28977983 Free PMC article. Review.
-
Identify latent chromosomal aberrations relevant to myelodysplastic syndromes.Sci Rep. 2017 Sep 4;7(1):10354. doi: 10.1038/s41598-017-10551-3. Sci Rep. 2017. PMID: 28871208 Free PMC article.
-
EZH2 mutations and promoter hypermethylation in childhood acute lymphoblastic leukemia.J Cancer Res Clin Oncol. 2016 Jul;142(7):1641-50. doi: 10.1007/s00432-016-2174-8. Epub 2016 May 11. J Cancer Res Clin Oncol. 2016. PMID: 27169594 Free PMC article.
-
Genomic variations in patients with myelodysplastic syndrome and karyotypes without numerical or structural changes.Sci Rep. 2021 Feb 2;11(1):2783. doi: 10.1038/s41598-021-81467-2. Sci Rep. 2021. PMID: 33531543 Free PMC article.
References
-
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. Genome-wide analysis of genetic alterations in acute lymphoblastic leukemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. - DOI - PubMed
-
- Suela J, Alvarez S, Cifuentes F, Largo C, Ferreira BI, Blesa D, Ardanaz M, Garcia R, Marquez JA, Odero MD, Calasanz MJ, Cigudosa JC. DNA profiling analysis of 100 consecutive de novo acute myeloid leukemia cases reveals patterns of genomic instability that affect all cytogenetic risk groups. Leukemia. 2007;21:1224–1231. doi: 10.1038/sj.leu.2404653. - DOI - PubMed
-
- Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, Yamatomo G, Nannya Y, Koehler R, Flohr T, Miller CW, Harbott J, Ludwig WD, Stanulla M, Schrappe M, Bartram CR, Koeffler HP. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high-resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2008;111(2):776–84. doi: 10.1182/blood-2007-05-088310. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

