Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase
- PMID: 25177796
- PMCID: PMC4209782
- DOI: 10.1042/BJ20140889
Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase
Abstract
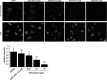
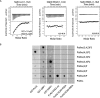
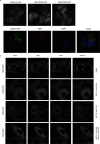
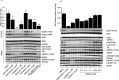
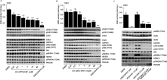
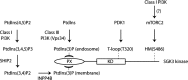
The Vps34 (vacuolar protein sorting 34) class III PI3K (phosphoinositide 3-kinase) phosphorylates PtdIns (phosphatidylinositol) at endosomal membranes to generate PtdIns(3)P that regulates membrane trafficking processes via its ability to recruit a subset of proteins possessing PtdIns(3)P-binding PX (phox homology) and FYVE domains. In the present study, we describe a highly selective and potent inhibitor of Vps34, termed VPS34-IN1, that inhibits Vps34 with 25 nM IC50 in vitro, but does not significantly inhibit the activity of 340 protein kinases or 25 lipid kinases tested that include all isoforms of class I as well as class II PI3Ks. Administration of VPS34-IN1 to cells induces a rapid dose-dependent dispersal of a specific PtdIns(3)P-binding probe from endosome membranes, within 1 min, without affecting the ability of class I PI3K to regulate Akt. Moreover, we explored whether SGK3 (serum- and glucocorticoid-regulated kinase-3), the only protein kinase known to interact specifically with PtdIns(3)P via its N-terminal PX domain, might be controlled by Vps34. Mutations disrupting PtdIns(3)P binding ablated SGK3 kinase activity by suppressing phosphorylation of the T-loop [PDK1 (phosphoinositide-dependent kinase 1) site] and hydrophobic motif (mammalian target of rapamycin site) residues. VPS34-IN1 induced a rapid ~50-60% loss of SGK3 phosphorylation within 1 min. VPS34-IN1 did not inhibit activity of the SGK2 isoform that does not possess a PtdIns(3)P-binding PX domain. Furthermore, class I PI3K inhibitors (GDC-0941 and BKM120) that do not inhibit Vps34 suppressed SGK3 activity by ~40%. Combining VPS34-IN1 and GDC-0941 reduced SGK3 activity ~80-90%. These data suggest SGK3 phosphorylation and hence activity is controlled by two pools of PtdIns(3)P. The first is produced through phosphorylation of PtdIns by Vps34 at the endosome. The second is due to the conversion of class I PI3K product, PtdIns(3,4,5)P3 into PtdIns(3)P, via the sequential actions of the PtdIns 5-phosphatases [SHIP1/2 (Src homology 2-domain-containing inositol phosphatase 1/2)] and PtdIns 4-phosphatase [INPP4B (inositol polyphosphate 4-phosphatase type II)]. VPS34-IN1 will be a useful probe to delineate physiological roles of the Vps34. Monitoring SGK3 phosphorylation and activity could be employed as a biomarker of Vps34 activity, in an analogous manner by which Akt is used to probe cellular class I PI3K activity. Combining class I (GDC-0941) and class III (VPS34-IN1) PI3K inhibitors could be used as a strategy to better analyse the roles and regulation of the elusive class II PI3K.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

