Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation
- PMID: 25180174
- PMCID: PMC4142390
- DOI: 10.1155/2014/970607
Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation
Abstract
Discovered in 1993, micoRNAs (miRNAs) are now recognized as one of the major regulatory gene families in eukaryotes. To date, 24521 microRNAs have been discovered and there are certainly more to come. It was primarily acknowledged that miRNAs result in gene expression repression at both the level of mRNA stability by conducting mRNA degradation and the level of translation (at initiation and after initiation) by inhibiting protein translation or degrading the polypeptides through binding complementarily to 3'UTR of the target mRNAs. Nevertheless, some studies revealed that miRNAs have the capability of activating gene expression directly or indirectly in respond to different cell types and conditions and in the presence of distinct cofactors. This reversibility in their posttranslational gene regulatory natures enables the bearing cells to rapidly response to different cell conditions and consequently block unnecessary energy wastage or maintain the cell state. This paper provides an overview of the current understandings of the miRNA characteristics including their genes and biogenesis, as well as their mediated downregulation. We also review up-to-date knowledge of miRNA-mediated gene upregulation through highlighting some notable examples and discuss the emerging concepts of their associations with other posttranscriptional gene regulation processes.
Figures

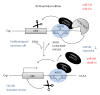

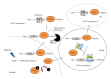
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. - PubMed
-
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology. 2009;10(2):126–139. - PubMed
-
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457(7228):396–404. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

