Using Pareto optimality to explore the topology and dynamics of the human connectome
- PMID: 25180308
- PMCID: PMC4150305
- DOI: 10.1098/rstb.2013.0530
Using Pareto optimality to explore the topology and dynamics of the human connectome
Abstract
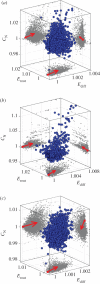
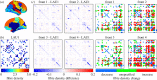
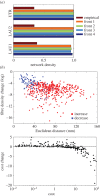
Graph theory has provided a key mathematical framework to analyse the architecture of human brain networks. This architecture embodies an inherently complex relationship between connection topology, the spatial arrangement of network elements, and the resulting network cost and functional performance. An exploration of these interacting factors and driving forces may reveal salient network features that are critically important for shaping and constraining the brain's topological organization and its evolvability. Several studies have pointed to an economic balance between network cost and network efficiency with networks organized in an 'economical' small-world favouring high communication efficiency at a low wiring cost. In this study, we define and explore a network morphospace in order to characterize different aspects of communication efficiency in human brain networks. Using a multi-objective evolutionary approach that approximates a Pareto-optimal set within the morphospace, we investigate the capacity of anatomical brain networks to evolve towards topologies that exhibit optimal information processing features while preserving network cost. This approach allows us to investigate network topologies that emerge under specific selection pressures, thus providing some insight into the selectional forces that may have shaped the network architecture of existing human brains.
Keywords: brain connectivity; diffusion imaging; graph theory; network science.
Figures


References
-
- Sporns O. 2011. Networks of the brain. Cambridge, MA: MIT Press.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

