Cyclin-dependent kinases
- PMID: 25180339
- PMCID: PMC4097832
- DOI: 10.1186/gb4184
Cyclin-dependent kinases
Abstract
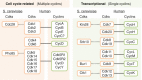
Cyclin-dependent kinases (CDKs) are protein kinases characterized by needing a separate subunit - a cyclin - that provides domains essential for enzymatic activity. CDKs play important roles in the control of cell division and modulate transcription in response to several extra- and intracellular cues. The evolutionary expansion of the CDK family in mammals led to the division of CDKs into three cell-cycle-related subfamilies (Cdk1, Cdk4 and Cdk5) and five transcriptional subfamilies (Cdk7, Cdk8, Cdk9, Cdk11 and Cdk20). Unlike the prototypical Cdc28 kinase of budding yeast, most of these CDKs bind one or a few cyclins, consistent with functional specialization during evolution. This review summarizes how, although CDKs are traditionally separated into cell-cycle or transcriptional CDKs, these activities are frequently combined in many family members. Not surprisingly, deregulation of this family of proteins is a hallmark of several diseases, including cancer, and drug-targeted inhibition of specific members has generated very encouraging results in clinical trials.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

