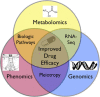
Improved drug therapy: triangulating phenomics with genomics and metabolomics
- PMID: 25181945
- PMCID: PMC4445687
- DOI: 10.1186/s40246-014-0016-9
Improved drug therapy: triangulating phenomics with genomics and metabolomics
Abstract
Embracing the complexity of biological systems has a greater likelihood to improve prediction of clinical drug response. Here we discuss limitations of a singular focus on genomics, epigenomics, proteomics, transcriptomics, metabolomics, or phenomics-highlighting the strengths and weaknesses of each individual technique. In contrast, 'systems biology' is proposed to allow clinicians and scientists to extract benefits from each technique, while limiting associated weaknesses by supplementing with other techniques when appropriate. Perfect predictive modeling is not possible, whereas modeling of intertwined phenomic responses using genomic stratification with metabolomic modifications may greatly improve predictive values for drug therapy. We thus propose a novel-integrated approach to personalized medicine that begins with phenomic data, is stratified by genomics, and ultimately refined by metabolomic pathway data. Whereas perfect prediction of efficacy and safety of drug therapy is not possible, improvements can be achieved by embracing the complexity of the biological system. Starting with phenomics, the combination of linking metabolomics to identify common biologic pathways and then stratifying by genomic architecture, might increase predictive values. This systems biology approach has the potential, in specific subsets of patients, to avoid drug therapy that will be either ineffective or unsafe.
Figures
References
-
- Ciaparrone M, Quirino M, Schinzari G, Zannoni G, Corsi DC, Vecchio FM, Cassano A, La Torre G, Barone C. Predictive role of thymidylate synthase, dihydropyrimidine dehydrogenase and thymidine phosphorylase expression in colorectal cancer patients receiving adjuvant 5-fluorouracil. Oncology. 2006;70:366–377. doi: 10.1159/000098110. - DOI - PubMed
-
- Lee A, Ezzeldin H, Fourie J, Diasio R. Dihydropyrimidine dehydrogenase deficiency: impact of pharmacogenetics on 5-fluorouracil therapy. Clin Adv Hematol Oncol. 2004;2:527–532. - PubMed
-
- Cappuzzo F, Finocchiaro G, Rossi E, Janne PA, Carnaghi C, Calandri C, Bencardino K, Ligorio C, Ciardiello F, Pressiani T, Destro A, Roncalli M, Crino L, Franklin WA, Santoro A, Varella-Garcia M. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19:717–723. doi: 10.1093/annonc/mdm492. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

