Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium
- PMID: 25182488
- PMCID: PMC4248824
- DOI: 10.1128/JB.02158-14
Patterns of expression and translocation of the ubiquitin ligase SlrP in Salmonella enterica serovar Typhimurium
Abstract
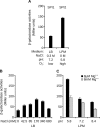
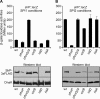
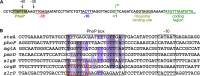
SlrP is an E3 ubiquitin ligase that can be translocated into eukaryotic host cells by the two type III secretion systems that are expressed by Salmonella enterica serovar Typhimurium and are encoded in Salmonella pathogenicity islands 1 (SPI1) and 2 (SPI2). Expression of slrP and translocation of its product were examined using lac, 3×FLAG, and cyaA' translational fusions. Although slrP was expressed in different media, optimal expression was found under conditions that imitate the intravacuolar environment and promote synthesis of the SPI2-encoded type III secretion system. Translocation into mammalian cells took place through the SPI1- or the SPI2-encoded type III secretion system, depending on specific host cell type and timing. A search for genetic factors involved in controlling the expression of slrP unveiled LeuO, Lon, and the two-component system PhoQ/PhoP as novel regulators of slrP. Our experiments suggest that LeuO and Lon act through HilD under SPI1-inducing conditions, whereas PhoP directly interacts with the slrP promoter to activate transcription under SPI2 inducing conditions.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
Specificities and redundancies in the NEL family of bacterial E3 ubiquitin ligases of Salmonella enterica serovar Typhimurium.Front Immunol. 2024 Feb 1;15:1328707. doi: 10.3389/fimmu.2024.1328707. eCollection 2024. Front Immunol. 2024. PMID: 38361917 Free PMC article.
-
Codependent and independent effects of nitric oxide-mediated suppression of PhoPQ and Salmonella pathogenicity island 2 on intracellular Salmonella enterica serovar typhimurium survival.Infect Immun. 2009 Nov;77(11):5107-15. doi: 10.1128/IAI.00759-09. Epub 2009 Sep 8. Infect Immun. 2009. PMID: 19737903 Free PMC article.
-
The Small RNA MicC Downregulates hilD Translation To Control the Salmonella Pathogenicity Island 1 Type III Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2022 Jan 18;204(1):e0037821. doi: 10.1128/JB.00378-21. Epub 2021 Oct 25. J Bacteriol. 2022. PMID: 34694902 Free PMC article.
-
PhoP-Mediated Repression of the SPI1 Type 3 Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2019 Jul 24;201(16):e00264-19. doi: 10.1128/JB.00264-19. Print 2019 Aug 15. J Bacteriol. 2019. PMID: 31182495 Free PMC article.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
Cited by
-
HilD induces expression of a novel Salmonella Typhimurium invasion factor, YobH, through a regulatory cascade involving SprB.Sci Rep. 2019 Sep 4;9(1):12725. doi: 10.1038/s41598-019-49192-z. Sci Rep. 2019. PMID: 31484980 Free PMC article.
-
How the PhoP/PhoQ System Controls Virulence and Mg2+ Homeostasis: Lessons in Signal Transduction, Pathogenesis, Physiology, and Evolution.Microbiol Mol Biol Rev. 2021 Aug 18;85(3):e0017620. doi: 10.1128/MMBR.00176-20. Epub 2021 Jun 30. Microbiol Mol Biol Rev. 2021. PMID: 34191587 Free PMC article. Review.
-
miR-215 Modulates Ubiquitination to Impair Inflammasome Activation and Autophagy During Salmonella Typhimurium Infection in Porcine Intestinal Cells.Animals (Basel). 2025 Feb 4;15(3):431. doi: 10.3390/ani15030431. Animals (Basel). 2025. PMID: 39943201 Free PMC article.
-
A Live Salmonella Vaccine Delivering PcrV through the Type III Secretion System Protects against Pseudomonas aeruginosa.mSphere. 2019 Apr 17;4(2):e00116-19. doi: 10.1128/mSphere.00116-19. mSphere. 2019. PMID: 30996108 Free PMC article.
-
The Hcp-like protein HilE inhibits homodimerization and DNA binding of the virulence-associated transcriptional regulator HilD in Salmonella.J Biol Chem. 2018 Apr 27;293(17):6578-6592. doi: 10.1074/jbc.RA117.001421. Epub 2018 Mar 13. J Biol Chem. 2018. PMID: 29535187 Free PMC article.
References
-
- Ramos-Morales F. 2012. Impact of Salmonella enterica type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol. 2012:787934. 10.5402/2012/787934. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

