Targeted gene suppression by inducing de novo DNA methylation in the gene promoter
- PMID: 25184003
- PMCID: PMC4150861
- DOI: 10.1186/1756-8935-7-20
Targeted gene suppression by inducing de novo DNA methylation in the gene promoter
Abstract
Background: Targeted gene silencing is an important approach in both drug development and basic research. However, the selection of a potent suppressor has become a significant hurdle to implementing maximal gene inhibition for this approach. We attempted to construct a 'super suppressor' by combining the activities of two suppressors that function through distinct epigenetic mechanisms.
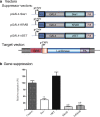
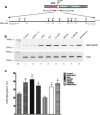
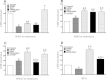
Results: Gene targeting vectors were constructed by fusing a GAL4 DNA-binding domain with a epigenetic suppressor, including CpG DNA methylase Sss1, histone H3 lysine 27 methylase vSET domain, and Kruppel-associated suppression box (KRAB). We found that both Sss1 and KRAB suppressors significantly inhibited the expression of luciferase and copGFP reporter genes. However, the histone H3 lysine 27 methylase vSET did not show significant suppression in this system. Constructs containing both Sss1 and KRAB showed better inhibition than either one alone. In addition, we show that KRAB suppressed gene expression by altering the histone code, but not DNA methylation in the gene promoter. Sss1, on the other hand, not only induced de novo DNA methylation and recruited Heterochromatin Protein 1 (HP1a), but also increased H3K27 and H3K9 methylation in the promoter.
Conclusions: Epigenetic studies can provide useful data for the selection of suppressors in constructing therapeutic vectors for targeted gene silencing.
Keywords: DNA methylation; Epigenetics; Gene expression; Gene suppression; H3K27 methylation; Histone code.
Figures


Similar articles
-
Epigenetic Targeting of Granulin in Hepatoma Cells by Synthetic CRISPR dCas9 Epi-suppressors.Mol Ther Nucleic Acids. 2018 Jun 1;11:23-33. doi: 10.1016/j.omtn.2018.01.002. Epub 2018 Jan 8. Mol Ther Nucleic Acids. 2018. PMID: 29858058 Free PMC article.
-
KRAB-Induced Heterochromatin Effectively Silences PLOD2 Gene Expression in Somatic Cells and is Resilient to TGFβ1 Activation.Int J Mol Sci. 2020 May 21;21(10):3634. doi: 10.3390/ijms21103634. Int J Mol Sci. 2020. PMID: 32455614 Free PMC article.
-
SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins.Genes Dev. 2002 Apr 15;16(8):919-32. doi: 10.1101/gad.973302. Genes Dev. 2002. PMID: 11959841 Free PMC article.
-
Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis.J Plant Physiol. 2006 Feb;163(3):358-68. doi: 10.1016/j.jplph.2005.10.015. Epub 2005 Dec 27. J Plant Physiol. 2006. PMID: 16384625 Review.
-
[Epigenetic regulation of histone H3 lysine 9 methylation in leukemia].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012 Feb;20(1):210-3. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2012. PMID: 22391199 Review. Chinese.
Cited by
-
Antitumor potential of a synthetic interferon-alpha/PLGF-2 positive charge peptide hybrid molecule in pancreatic cancer cells.Sci Rep. 2015 Nov 20;5:16975. doi: 10.1038/srep16975. Sci Rep. 2015. PMID: 26584517 Free PMC article.
-
The expression signature of cancer-associated KRAB-ZNF factors identified in TCGA pan-cancer transcriptomic data.Mol Oncol. 2019 Apr;13(4):701-724. doi: 10.1002/1878-0261.12407. Epub 2019 Feb 16. Mol Oncol. 2019. PMID: 30444046 Free PMC article.
-
Molecular structures guide the engineering of chromatin.Nucleic Acids Res. 2017 Jul 27;45(13):7555-7570. doi: 10.1093/nar/gkx531. Nucleic Acids Res. 2017. PMID: 28609787 Free PMC article.
-
Oplr16 serves as a novel chromatin factor to control stem cell fate by modulating pluripotency-specific chromosomal looping and TET2-mediated DNA demethylation.Nucleic Acids Res. 2020 Apr 17;48(7):3935-3948. doi: 10.1093/nar/gkaa097. Nucleic Acids Res. 2020. PMID: 32055844 Free PMC article.
-
Human cytomegalovirus hijacks host stress response fueling replication stress and genome instability.Cell Death Differ. 2022 Aug;29(8):1639-1653. doi: 10.1038/s41418-022-00953-w. Epub 2022 Feb 22. Cell Death Differ. 2022. PMID: 35194187 Free PMC article.
References
-
- Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–11928. - PubMed
-
- Murad JP, Lin OA, Espinosa EV, Khasawneh FT. Current and experimental antibody-based therapeutics: insights, breakthroughs, setbacks and future directions. Curr Mol Med. 2013;13:165–178. - PubMed
-
- Sapra P, Shor B. Monoclonal antibody-based therapies in cancer: advances and challenges. Pharmacol Ther. 2013;138:452–469. - PubMed
-
- Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

