DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum
- PMID: 25184562
- PMCID: PMC4187680
- DOI: 10.1128/MMBR.00059-13
DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum
Abstract
Research into the complex genetic underpinnings of the malaria parasite Plasmodium falciparum is entering a new era with the arrival of site-specific genome engineering. Previously restricted only to model systems but now expanded to most laboratory organisms, and even to humans for experimental gene therapy studies, this technology allows researchers to rapidly generate previously unattainable genetic modifications. This technological advance is dependent on DNA double-strand break repair (DSBR), specifically homologous recombination in the case of Plasmodium. Our understanding of DSBR in malaria parasites, however, is based largely on assumptions and knowledge taken from other model systems, which do not always hold true in Plasmodium. Here we describe the causes of double-strand breaks, the mechanisms of DSBR, and the differences between model systems and P. falciparum. These mechanisms drive basic parasite functions, such as meiosis, antigen diversification, and copy number variation, and allow the parasite to continually evolve in the contexts of host immune pressure and drug selection. Finally, we discuss the new technologies that leverage DSBR mechanisms to accelerate genetic investigations into this global infectious pathogen.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



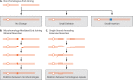
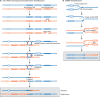
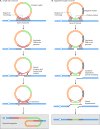



Similar articles
-
Rapid antigen diversification through mitotic recombination in the human malaria parasite Plasmodium falciparum.PLoS Biol. 2019 May 13;17(5):e3000271. doi: 10.1371/journal.pbio.3000271. eCollection 2019 May. PLoS Biol. 2019. PMID: 31083650 Free PMC article.
-
The contribution of extrachromosomal DNA to genome plasticity in malaria parasites.Mol Microbiol. 2021 Apr;115(4):503-507. doi: 10.1111/mmi.14632. Epub 2020 Nov 19. Mol Microbiol. 2021. PMID: 33103309 Free PMC article.
-
Genome sequence of the human malaria parasite Plasmodium falciparum.Nature. 2002 Oct 3;419(6906):498-511. doi: 10.1038/nature01097. Nature. 2002. PMID: 12368864 Free PMC article.
-
The selection landscape of malaria parasites.Science. 2010 May 14;328(5980):866-71. doi: 10.1126/science.1185410. Science. 2010. PMID: 20466925 Review.
-
Genetic diversity and population history of Plasmodium falciparum and Plasmodium vivax.Parassitologia. 2006 Dec;48(4):561-6. Parassitologia. 2006. PMID: 17688177 Review. No abstract available.
Cited by
-
Identification of Plasmodium falciparum DNA Repair Protein Mre11 with an Evolutionarily Conserved Nuclease Function.PLoS One. 2015 May 4;10(5):e0125358. doi: 10.1371/journal.pone.0125358. eCollection 2015. PLoS One. 2015. PMID: 25938776 Free PMC article.
-
Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies.Nat Commun. 2016 May 18;7:11553. doi: 10.1038/ncomms11553. Nat Commun. 2016. PMID: 27189525 Free PMC article.
-
CRISPR/Cas9 Editing of the Plasmodium falciparum Genome.Methods Mol Biol. 2022;2470:221-239. doi: 10.1007/978-1-0716-2189-9_17. Methods Mol Biol. 2022. PMID: 35881349 Review.
-
Comparative Analysis of Plasmodium falciparum Genotyping via SNP Detection, Microsatellite Profiling, and Whole-Genome Sequencing.Antimicrob Agents Chemother. 2022 Jan 18;66(1):e0116321. doi: 10.1128/AAC.01163-21. Epub 2021 Oct 25. Antimicrob Agents Chemother. 2022. PMID: 34694871 Free PMC article.
-
CRISPR-mediated genome editing of Plasmodium falciparum malaria parasites.Genome Med. 2014 Aug 26;6(8):63. doi: 10.1186/s13073-014-0063-9. eCollection 2014. Genome Med. 2014. PMID: 25473431 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

