Transcript-RNA-templated DNA recombination and repair
- PMID: 25186730
- PMCID: PMC4899968
- DOI: 10.1038/nature13682
Transcript-RNA-templated DNA recombination and repair
Abstract
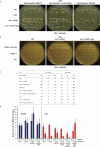
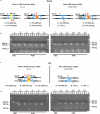
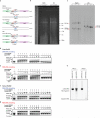
Homologous recombination is a molecular process that has multiple important roles in DNA metabolism, both for DNA repair and genetic variation in all forms of life. Generally, homologous recombination involves the exchange of genetic information between two identical or nearly identical DNA molecules; however, homologous recombination can also occur between RNA molecules, as shown for RNA viruses. Previous research showed that synthetic RNA oligonucleotides can act as templates for DNA double-strand break (DSB) repair in yeast and human cells, and artificial long RNA templates injected in ciliate cells can guide genomic rearrangements. Here we report that endogenous transcript RNA mediates homologous recombination with chromosomal DNA in yeast Saccharomyces cerevisiae. We developed a system to detect the events of homologous recombination initiated by transcript RNA following the repair of a chromosomal DSB occurring either in a homologous but remote locus, or in the same transcript-generating locus in reverse-transcription-defective yeast strains. We found that RNA-DNA recombination is blocked by ribonucleases H1 and H2. In the presence of H-type ribonucleases, DSB repair proceeds through a complementary DNA intermediate, whereas in their absence, it proceeds directly through RNA. The proximity of the transcript to its chromosomal DNA partner in the same locus facilitates Rad52-driven homologous recombination during DSB repair. We demonstrate that yeast and human Rad52 proteins efficiently catalyse annealing of RNA to a DSB-like DNA end in vitro. Our results reveal a novel mechanism of homologous recombination and DNA repair in which transcript RNA is used as a template for DSB repair. Thus, considering the abundance of RNA transcripts in cells, RNA may have a marked impact on genomic stability and plasticity.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

