Functional genomics of lactic acid bacteria: from food to health
- PMID: 25186768
- PMCID: PMC4155825
- DOI: 10.1186/1475-2859-13-S1-S8
Functional genomics of lactic acid bacteria: from food to health
Abstract
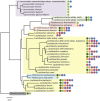
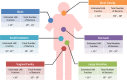
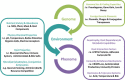
Genome analysis using next generation sequencing technologies has revolutionized the characterization of lactic acid bacteria and complete genomes of all major groups are now available. Comparative genomics has provided new insights into the natural and laboratory evolution of lactic acid bacteria and their environmental interactions. Moreover, functional genomics approaches have been used to understand the response of lactic acid bacteria to their environment. The results have been instrumental in understanding the adaptation of lactic acid bacteria in artisanal and industrial food fermentations as well as their interactions with the human host. Collectively, this has led to a detailed analysis of genes involved in colonization, persistence, interaction and signaling towards to the human host and its health. Finally, massive parallel genome re-sequencing has provided new opportunities in applied genomics, specifically in the characterization of novel non-GMO strains that have potential to be used in the food industry. Here, we provide an overview of the state of the art of these functional genomics approaches and their impact in understanding, applying and designing lactic acid bacteria for food and health.
Figures
Similar articles
-
Comparative genomics of the lactic acid bacteria.Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15611-6. doi: 10.1073/pnas.0607117103. Epub 2006 Oct 9. Proc Natl Acad Sci U S A. 2006. PMID: 17030793 Free PMC article.
-
Genomics of lactic acid bacteria.FEMS Microbiol Lett. 2009 Mar;292(1):1-6. doi: 10.1111/j.1574-6968.2008.01442.x. Epub 2008 Dec 11. FEMS Microbiol Lett. 2009. PMID: 19087207 Review.
-
[Microevolution of lactic acid bacteria--A review].Wei Sheng Wu Xue Bao. 2015 Nov 4;55(11):1371-7. Wei Sheng Wu Xue Bao. 2015. PMID: 26915217 Review. Chinese.
-
Comparative genomics of enzymes in flavor-forming pathways from amino acids in lactic acid bacteria.Appl Environ Microbiol. 2008 Aug;74(15):4590-600. doi: 10.1128/AEM.00150-08. Epub 2008 Jun 6. Appl Environ Microbiol. 2008. PMID: 18539796 Free PMC article. Review. No abstract available.
-
Convergent reductive evolution in bee-associated lactic acid bacteria.Appl Environ Microbiol. 2024 Nov 20;90(11):e0125724. doi: 10.1128/aem.01257-24. Epub 2024 Oct 23. Appl Environ Microbiol. 2024. PMID: 39440949 Free PMC article.
Cited by
-
Phylotype-Level Characterization of Complex Communities of Lactobacilli Using a High-Throughput, High-Resolution Phenylalanyl-tRNA Synthetase (pheS) Gene Amplicon Sequencing Approach.Appl Environ Microbiol. 2020 Dec 17;87(1):e02191-20. doi: 10.1128/AEM.02191-20. Print 2020 Dec 17. Appl Environ Microbiol. 2020. PMID: 33097506 Free PMC article.
-
Dynamic and Functional Characteristics of Predominant Species in Industrial Paocai as Revealed by Combined DGGE and Metagenomic Sequencing.Front Microbiol. 2018 Oct 9;9:2416. doi: 10.3389/fmicb.2018.02416. eCollection 2018. Front Microbiol. 2018. PMID: 30356774 Free PMC article.
-
Comparison of 16S rRNA Gene Based Microbial Profiling Using Five Next-Generation Sequencers and Various Primers.Front Microbiol. 2021 Oct 14;12:715500. doi: 10.3389/fmicb.2021.715500. eCollection 2021. Front Microbiol. 2021. PMID: 34721319 Free PMC article.
-
Engineered probiotic Lactobacillus plantarum WCSF I for monitoring and treatment of Staphylococcus aureus infection.Microbiol Spectr. 2023 Dec 12;11(6):e0182923. doi: 10.1128/spectrum.01829-23. Epub 2023 Nov 1. Microbiol Spectr. 2023. PMID: 37909791 Free PMC article.
-
Characterization and Safety Evaluation of Autoclaved Gut Commensal Parabacteroides goldsteinii RV-01.Int J Mol Sci. 2024 Nov 25;25(23):12660. doi: 10.3390/ijms252312660. Int J Mol Sci. 2024. PMID: 39684372 Free PMC article.
References
-
- Tannock GW. Normal microflora: an introduction to microbes inhabiting the human body. Springer; 1995.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

