Image-based high-throughput field phenotyping of crop roots
- PMID: 25187526
- PMCID: PMC4213080
- DOI: 10.1104/pp.114.243519
Image-based high-throughput field phenotyping of crop roots
Abstract
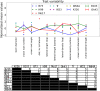
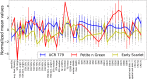
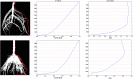
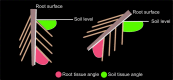
Current plant phenotyping technologies to characterize agriculturally relevant traits have been primarily developed for use in laboratory and/or greenhouse conditions. In the case of root architectural traits, this limits phenotyping efforts, largely, to young plants grown in specialized containers and growth media. Hence, novel approaches are required to characterize mature root systems of older plants grown under actual soil conditions in the field. Imaging methods able to address the challenges associated with characterizing mature root systems are rare due, in part, to the greater complexity of mature root systems, including the larger size, overlap, and diversity of root components. Our imaging solution combines a field-imaging protocol and algorithmic approach to analyze mature root systems grown in the field. Via two case studies, we demonstrate how image analysis can be utilized to estimate localized root traits that reliably capture heritable architectural diversity as well as environmentally induced architectural variation of both monocot and dicot plants. In the first study, we show that our algorithms and traits (including 13 novel traits inaccessible to manual estimation) can differentiate nine maize (Zea mays) genotypes 8 weeks after planting. The second study focuses on a diversity panel of 188 cowpea (Vigna unguiculata) genotypes to identify which traits are sufficient to differentiate genotypes even when comparing plants whose harvesting date differs up to 14 d. Overall, we find that automatically derived traits can increase both the speed and reproducibility of the trait estimation pipeline under field conditions.
© 2014 American Society of Plant Biologists. All Rights Reserved.
Figures








References
-
- Araus JL, Cairns JE. (2014) Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci 19: 52–61 - PubMed
-
- Armengaud P, Zambaux K, Hills A, Sulpice R, Pattison RJ, Blatt MR, Amtmann A. (2009) EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J 57: 945–956 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

