Population structure of clinical Pseudomonas aeruginosa from West and Central African countries
- PMID: 25187957
- PMCID: PMC4154784
- DOI: 10.1371/journal.pone.0107008
Population structure of clinical Pseudomonas aeruginosa from West and Central African countries
Abstract
Background: Pseudomonas aeruginosa (PA) has a non-clonal, epidemic population with a few widely distributed and frequently encountered sequence types (STs) called 'high-risk clusters'. Clinical P. aeruginosa (clinPA) has been studied in all inhabited continents excepted in Africa, where a very few isolates have been analyzed. Here, we characterized a collection of clinPA isolates from four countries of West and Central Africa.
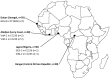
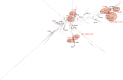
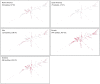
Methodology: 184 non-redundant isolates of clinPA from hospitals of Senegal, Ivory Coast, Nigeria, and Central African Republic were genotyped by MLST. We assessed their resistance level to antibiotics by agar diffusion and identified the extended-spectrum β-lactamases (ESBLs) and metallo-β-lactamases (MBLs) by sequencing. The population structure of the species was determined by a nucleotide-based analysis of the entire PA MLST database and further localized on the phylogenetic tree (i) the sequence types (STs) of the present collection, (ii) the STs by continents, (iii) ESBL- and MBL-producing STs from the MLST database.
Principal findings: We found 80 distinct STs, of which 24 had no relationship with any known STs. 'High-risk' international clonal complexes (CC155, CC244, CC235) were frequently found in West and Central Africa. The five VIM-2-producing isolates belonged to CC233 and CC244. GES-1 and GES-9 enzymes were produced by one CC235 and one ST1469 isolate, respectively. We showed the spread of 'high-risk' international clonal complexes, often described as multidrug-resistant on other continents, with a fully susceptible phenotype. The MBL- and ESBL-producing STs were scattered throughout the phylogenetic tree and our data suggest a poor association between a continent and a specific phylogroup.
Conclusions: ESBL- and MBL-encoding genes are borne by both successful international clonal complexes and distinct local STs in clinPA of West and Central Africa. Furthermore, our data suggest that the spread of a ST could be either due to its antibiotic resistance or to features independent from the resistance to antibiotics.
Conflict of interest statement
Figures

References
-
- Lyczak JB, Cannon CL, Pier GB (2000) Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect 2: 1051–1060. - PubMed
-
- Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, et al. (2007) Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13: 560–578. - PubMed
-
- Breidenstein EBM, de la Fuente-Núñez C, Hancock REW (2011) Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19: 419–426. - PubMed
-
- Woodford N, Turton JF, Livermore DM (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35: 736–755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

