Viral RNAs are unusually compact
- PMID: 25188030
- PMCID: PMC4154850
- DOI: 10.1371/journal.pone.0105875
Viral RNAs are unusually compact
Abstract
A majority of viruses are composed of long single-stranded genomic RNA molecules encapsulated by protein shells with diameters of just a few tens of nanometers. We examine the extent to which these viral RNAs have evolved to be physically compact molecules to facilitate encapsulation. Measurements of equal-length viral, non-viral, coding and non-coding RNAs show viral RNAs to have among the smallest sizes in solution, i.e., the highest gel-electrophoretic mobilities and the smallest hydrodynamic radii. Using graph-theoretical analyses we demonstrate that their sizes correlate with the compactness of branching patterns in predicted secondary structure ensembles. The density of branching is determined by the number and relative positions of 3-helix junctions, and is highly sensitive to the presence of rare higher-order junctions with 4 or more helices. Compact branching arises from a preponderance of base pairing between nucleotides close to each other in the primary sequence. The density of branching represents a degree of freedom optimized by viral RNA genomes in response to the evolutionary pressure to be packaged reliably. Several families of viruses are analyzed to delineate the effects of capsid geometry, size and charge stabilization on the selective pressure for RNA compactness. Compact branching has important implications for RNA folding and viral assembly.
Conflict of interest statement
Figures

 molecules. B3 & Y2 were mixed prior to running in lane 6. Mobility is measured as the distance from the DNA marker (see Methods), and reported relative to B3.
molecules. B3 & Y2 were mixed prior to running in lane 6. Mobility is measured as the distance from the DNA marker (see Methods), and reported relative to B3.
 , (C) tree-graph radii of gyration Rg, (D) higher-order branching propensity
, (C) tree-graph radii of gyration Rg, (D) higher-order branching propensity  , and (E) numbers of d = 4 (circles) and d≥4 (squares) vertices. Solid lines are least-squares linear regression fits. Error bars are standard deviations (
, and (E) numbers of d = 4 (circles) and d≥4 (squares) vertices. Solid lines are least-squares linear regression fits. Error bars are standard deviations ( ) except in A, where they are the standard errors of estimates (
) except in A, where they are the standard errors of estimates ( ). Standard deviations of
). Standard deviations of  are listed in Table S1 in File S1.
are listed in Table S1 in File S1.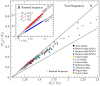
 is shown versus
is shown versus  in both plots. Inset B shows 4000-nt random-sequence data (gray squares) with
in both plots. Inset B shows 4000-nt random-sequence data (gray squares) with  (red squares) and
(red squares) and  (blue squares) plotted against
(blue squares) plotted against  (see Eqs. 4 & 5). Values of
(see Eqs. 4 & 5). Values of  /
/ (gray squares) are consistent with
(gray squares) are consistent with  , indicating that most higher-order junctions in random RNAs have
, indicating that most higher-order junctions in random RNAs have  . Plot A compares the random sequences with eleven distinct families of viral RNA. Families with more than half their members having
. Plot A compares the random sequences with eleven distinct families of viral RNA. Families with more than half their members having  are shown with circular symbols.
are shown with circular symbols.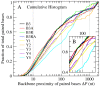
 ) are in F & G. Viral and non-viral histograms diverge up to
) are in F & G. Viral and non-viral histograms diverge up to  and converge thereafter. Unlike in yeast RNAs, over 70% of base pairs in B3 (See inset G) have
and converge thereafter. Unlike in yeast RNAs, over 70% of base pairs in B3 (See inset G) have  . This predominance of proximal base pairing leads to compact secondary structures.
. This predominance of proximal base pairing leads to compact secondary structures.References
-
- Tinoco I, Bustamante C (1999) How RNA folds. J Mol Biol 293: 271–281. - PubMed
-
- Li PTX, Vieregg J, Tinoco I (2008) How RNA unfolds and refolds. Annu Rev Biochem 77: 77–100. - PubMed
-
- Mathews DH, Turner DH (2006) Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 16: 270–278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

