Pronounced metabolic changes in adaptation to biofilm growth by Streptococcus pneumoniae
- PMID: 25188255
- PMCID: PMC4154835
- DOI: 10.1371/journal.pone.0107015
Pronounced metabolic changes in adaptation to biofilm growth by Streptococcus pneumoniae
Abstract
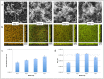
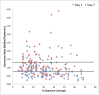
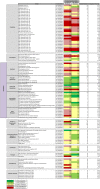
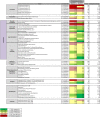
Streptococcus pneumoniae accounts for a significant global burden of morbidity and mortality and biofilm development is increasingly recognised as important for colonization and infection. Analysis of protein expression patterns during biofilm development may therefore provide valuable insights to the understanding of pneumococcal persistence strategies and to improve vaccines. iTRAQ (isobaric tagging for relative and absolute quantification), a high-throughput gel-free proteomic approach which allows high resolution quantitative comparisons of protein profiles between multiple phenotypes, was used to interrogate planktonic and biofilm growth in a clinical serotype 14 strain. Comparative analyses of protein expression between log-phase planktonic and 1-day and 7-day biofilm cultures representing nascent and late phase biofilm growth were carried out. Overall, 244 proteins were identified, of which >80% were differentially expressed during biofilm development. Quantitatively and qualitatively, metabolic regulation appeared to play a central role in the adaptation from the planktonic to biofilm phenotype. Pneumococci adapted to biofilm growth by decreasing enzymes involved in the glycolytic pathway, as well as proteins involved in translation, transcription, and virulence. In contrast, proteins with a role in pyruvate, carbohydrate, and arginine metabolism were significantly increased during biofilm development. Downregulation of glycolytic and translational proteins suggests that pneumococcus adopts a covert phenotype whilst adapting to an adherent lifestyle, while utilization of alternative metabolic pathways highlights the resourcefulness of pneumococcus to facilitate survival in diverse environmental conditions. These metabolic proteins, conserved across both the planktonic and biofilm phenotypes, may also represent target candidates for future vaccine development and treatment strategies. Data are available via ProteomeXchange with identifier PXD001182.
Conflict of interest statement
Figures
References
-
- Kadioglu A, Weiser JN, Paton JC, Andrew PW (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6: 288–301. - PubMed
-
- Reid SD, Hong W, Dew KE, Winn DR, Pang B, et al. (2009) Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J Infect Dis 199: 786–794. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

