The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7
- PMID: 25188390
- PMCID: PMC4154866
- DOI: 10.1371/journal.ppat.1004331
The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7
Erratum in
-
Correction: The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7.PLoS Pathog. 2015 Feb 19;11(2):e1004710. doi: 10.1371/journal.ppat.1004710. eCollection 2015 Feb. PLoS Pathog. 2015. PMID: 25695800 Free PMC article.
Abstract
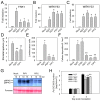
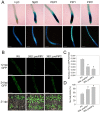
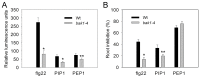
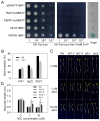
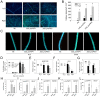
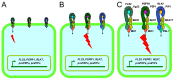
In plants, innate immune responses are initiated by plasma membrane-located pattern recognition receptors (PRRs) upon recognition of elicitors, including exogenous pathogen-associated molecular patterns (PAMPs) and endogenous damage-associated molecular patterns (DAMPs). Arabidopsis thaliana produces more than 1000 secreted peptide candidates, but it has yet to be established whether any of these act as elicitors. Here we identified an A. thaliana gene family encoding precursors of PAMP-induced secreted peptides (prePIPs) through an in-silico approach. The expression of some members of the family, including prePIP1 and prePIP2, is induced by a variety of pathogens and elicitors. Subcellular localization and proteolytic processing analyses demonstrated that the prePIP1 product is secreted into extracellular spaces where it is cleaved at the C-terminus. Overexpression of prePIP1 and prePIP2, or exogenous application of PIP1 and PIP2 synthetic peptides corresponding to the C-terminal conserved regions in prePIP1 and prePIP2, enhanced immune responses and pathogen resistance in A. thaliana. Genetic and biochemical analyses suggested that the receptor-like kinase 7 (RLK7) functions as a receptor of PIP1. Once perceived by RLK7, PIP1 initiates overlapping and distinct immune signaling responses together with the DAMP PEP1. PIP1 and PEP1 cooperate in amplifying the immune responses triggered by the PAMP flagellin. Collectively, these studies provide significant insights into immune modulation by Arabidopsis endogenous secreted peptides.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
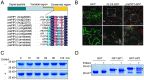
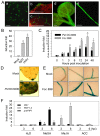
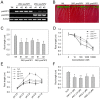
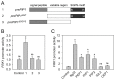

References
-
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329. - PubMed
-
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. - PubMed
-
- Sanabria N, Goring D, Nurnberger T, Dubery I (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self-incompatibility and innate immunity. New Phytol 178: 503–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

