Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA
- PMID: 25188500
- PMCID: PMC4154765
- DOI: 10.1371/journal.pone.0107000
Crystal structure of the mineralocorticoid receptor DNA binding domain in complex with DNA
Abstract
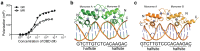
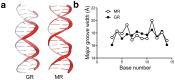
The steroid hormone receptors regulate important physiological functions such as reproduction, metabolism, immunity, and electrolyte balance. Mutations within steroid receptors result in endocrine disorders and can often drive cancer formation and progression. Despite the conserved three-dimensional structure shared among members of the steroid receptor family and their overlapping DNA binding preference, activation of individual steroid receptors drive unique effects on gene expression. Here, we present the first structure of the human mineralocorticoid receptor DNA binding domain, in complex with a canonical DNA response element. The overall structure is similar to the glucocorticoid receptor DNA binding domain, but small changes in the mode of DNA binding and lever arm conformation may begin to explain the differential effects on gene regulation by the mineralocorticoid and glucocorticoid receptors. In addition, we explore the structural effects of mineralocorticoid receptor DNA binding domain mutations found in type I pseudohypoaldosteronism and multiple types of cancer.
Conflict of interest statement
Figures
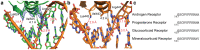




References
-
- Falkenstein E, Tillmann HC, Christ M, Feuring M, Wehling M (2000) Multiple actions of steroid hormones–a focus on rapid, nongenomic effects. Pharmacol Rev 52: 513–556. - PubMed
-
- Tsai M, O’Malley BW (1994) Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem 63: 451–486. - PubMed
-
- Bridgham JT, Carroll SM, Thornton JW (2006) Evolution of hormone-receptor complexity by molecular exploitation. Science 312: 97–101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

