Regulation of DNA pairing in homologous recombination
- PMID: 25190078
- PMCID: PMC4413238
- DOI: 10.1101/cshperspect.a017954
Regulation of DNA pairing in homologous recombination
Abstract
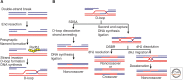
Homologous recombination (HR) is a major mechanism for eliminating DNA double-strand breaks from chromosomes. In this process, the break termini are resected nucleolytically to form 3' ssDNA (single-strand DNA) overhangs. A recombinase (i.e., a protein that catalyzes homologous DNA pairing and strand exchange) assembles onto the ssDNA and promotes pairing with a homologous duplex. DNA synthesis then initiates from the 3' end of the invading strand, and the extended DNA joint is resolved via one of several pathways to restore the integrity of the injured chromosome. It is crucial that HR be carefully orchestrated because spurious events can create cytotoxic intermediates or cause genomic rearrangements and loss of gene heterozygosity, which can lead to cell death or contribute to the development of cancer. In this review, we will discuss how DNA motor proteins regulate HR via a dynamic balance of the recombination-promoting and -attenuating activities that they possess.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
-
- Alexeev A, Mazin A, Kowalczykowski SC 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol 10: 182–186. - PubMed
-
- Bartram CR, Koske-Westphal T, Passarge E 1976. Chromatid exchanges in ataxia telangiectasia, Bloom syndrome, Werner syndrome, and xeroderma pigmentosum. Ann Hum Genet 40: 79–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
