Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours
- PMID: 25190313
- PMCID: PMC4159681
- DOI: 10.1038/ncomms5802
Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours
Erratum in
-
Erratum: Publisher Correction: Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours.Nat Commun. 2017 Nov 29;8:16177. doi: 10.1038/ncomms16177. eCollection 2017. Nat Commun. 2017. PMID: 31305780 Free PMC article.
Abstract
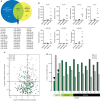
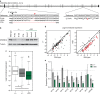
Wilms tumour is the most common childhood kidney cancer. Here we report the whole-exome sequencing of 44 Wilms tumours, identifying missense mutations in the microRNA (miRNA)-processing enzymes DROSHA and DICER1, and novel mutations in MYCN, SMARCA4 and ARID1A. Examination of tumour miRNA expression, in vitro processing assays and genomic editing in human cells demonstrates that DICER1 and DROSHA mutations influence miRNA processing through distinct mechanisms. DICER1 RNase IIIB mutations preferentially impair processing of miRNAs deriving from the 5'-arm of pre-miRNA hairpins, while DROSHA RNase IIIB mutations globally inhibit miRNA biogenesis through a dominant-negative mechanism. Both DROSHA and DICER1 mutations impair expression of tumour-suppressing miRNAs, including the let-7 family, important regulators of MYCN, LIN28 and other Wilms tumour oncogenes. These results provide new insights into the mechanisms through which mutations in miRNA biogenesis components reprogramme miRNA expression in human cancer and suggest that these defects define a distinct subclass of Wilms tumours.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Chu A. et al.. Wilms’ tumour: a systematic review of risk factors and meta-analysis. Paediatr. Perinat. Epidemiol. 24, 449–469 (2010). - PubMed
-
- Rivera M. N. & Haber D. A. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat. Rev. Cancer 5, 699–712 (2005). - PubMed
-
- Ko E. Y. & Ritchey M. L. Current management of Wilms’ tumor in children. J. Pediatr. Urol. 5, 56–65 (2009). - PubMed
-
- Kalapurakal J. A. et al.. Management of Wilms’ tumour: current practice and future goals. Lancet Oncol. 5, 37–46 (2004). - PubMed
-
- Murphy W. M., Grignon D. J. & Perlman E. J. inAFIP Atlas of Tumor Pathology Series 4. Tumors of the Kidney, Bladder, and Related Urinary Structures ed. Silverberg S. G. American Registry of Pathology (2004).
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

