Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells
- PMID: 25190717
- PMCID: PMC4390554
- DOI: 10.1126/science.1256888
Direct roles of SPEECHLESS in the specification of stomatal self-renewing cells
Abstract
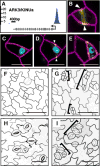
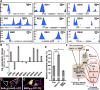
Lineage-specific stem cells are critical for the production and maintenance of specific cell types and tissues in multicellular organisms. In Arabidopsis, the initiation and proliferation of stomatal lineage cells is controlled by the basic helix-loop-helix transcription factor SPEECHLESS (SPCH). SPCH-driven asymmetric and self-renewing divisions allow flexibility in stomatal production and overall organ growth. How SPCH directs stomatal lineage cell behaviors, however, is unclear. Here, we improved the chromatin immunoprecipitation (ChIP) assay and profiled the genome-wide targets of Arabidopsis SPCH in vivo. We found that SPCH controls key regulators of cell fate and asymmetric cell divisions and modulates responsiveness to peptide and phytohormone-mediated intercellular communication. Our results delineate the molecular pathways that regulate an essential adult stem cell lineage in plants.
Copyright © 2014, American Association for the Advancement of Science.
Figures
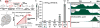

Similar articles
-
Modulation of Asymmetric Division Diversity through Cytokinin and SPEECHLESS Regulatory Interactions in the Arabidopsis Stomatal Lineage.Dev Cell. 2018 Oct 8;47(1):53-66.e5. doi: 10.1016/j.devcel.2018.08.007. Epub 2018 Sep 6. Dev Cell. 2018. PMID: 30197241 Free PMC article.
-
SCREAM promotes the timely termination of proliferative divisions in stomatal lineage by directly suppressing SPEECHLESS transcription.New Phytol. 2025 Jun;246(6):2565-2579. doi: 10.1111/nph.70145. Epub 2025 Apr 24. New Phytol. 2025. PMID: 40275539
-
Phosphorylation of Serine 186 of bHLH Transcription Factor SPEECHLESS Promotes Stomatal Development in Arabidopsis.Mol Plant. 2015 May;8(5):783-95. doi: 10.1016/j.molp.2014.12.014. Epub 2014 Dec 30. Mol Plant. 2015. PMID: 25680231
-
The plant stomatal lineage at a glance.J Cell Sci. 2019 Apr 26;132(8):jcs228551. doi: 10.1242/jcs.228551. J Cell Sci. 2019. PMID: 31028153 Free PMC article. Review.
-
Transcriptional control of cell fate in the stomatal lineage.Curr Opin Plant Biol. 2016 Feb;29:1-8. doi: 10.1016/j.pbi.2015.09.008. Epub 2015 Nov 7. Curr Opin Plant Biol. 2016. PMID: 26550955 Free PMC article. Review.
Cited by
-
Stomatal development in the changing climate.Development. 2024 Oct 15;151(20):dev202681. doi: 10.1242/dev.202681. Epub 2024 Oct 21. Development. 2024. PMID: 39431330 Free PMC article. Review.
-
Stomagenesis versus myogenesis: Parallels in intrinsic and extrinsic regulation of transcription factor mediated specialized cell-type differentiation in plants and animals.Dev Growth Differ. 2016 May;58(4):341-54. doi: 10.1111/dgd.12282. Epub 2016 Apr 29. Dev Growth Differ. 2016. PMID: 27125444 Free PMC article. Review.
-
Cell-type-specific transcriptome and histone modification dynamics during cellular reprogramming in the Arabidopsis stomatal lineage.Proc Natl Acad Sci U S A. 2019 Oct 22;116(43):21914-21924. doi: 10.1073/pnas.1911400116. Epub 2019 Oct 8. Proc Natl Acad Sci U S A. 2019. PMID: 31594845 Free PMC article.
-
A Mutation in the bHLH Domain of the SPCH Transcription Factor Uncovers a BR-Dependent Mechanism for Stomatal Development.Plant Physiol. 2017 Jun;174(2):823-842. doi: 10.1104/pp.17.00615. Epub 2017 May 15. Plant Physiol. 2017. PMID: 28507175 Free PMC article.
-
LLM-Domain B-GATA Transcription Factors Promote Stomatal Development Downstream of Light Signaling Pathways in Arabidopsis thaliana Hypocotyls.Plant Cell. 2016 Mar;28(3):646-60. doi: 10.1105/tpc.15.00783. Epub 2016 Feb 25. Plant Cell. 2016. PMID: 26917680 Free PMC article.
References
-
- Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 2014;15:301–312. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

