An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes
- PMID: 25191222
- PMCID: PMC4137239
- DOI: 10.3389/fnmol.2014.00072
An optimal ubiquitin-proteasome pathway in the nervous system: the role of deubiquitinating enzymes
Abstract
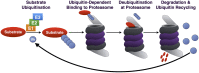
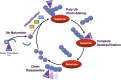
The Ubiquitin-Proteasome Pathway (UPP), which is critical for normal function in the nervous system and is implicated in various neurological diseases, requires the small modifier protein ubiquitin to accomplish its duty of selectively degrading short-lived, abnormal or misfolded proteins. Over the past decade, a large class of proteases collectively known as deubiquitinating enzymes (DUBs) has increasingly gained attention in all manners related to ubiquitin. By cleaving ubiquitin from another protein, DUBs ensure that the UPP functions properly. DUBs accomplish this task by processing newly translated ubiquitin so that it can be used for conjugation to substrate proteins, by regulating the "where, when, and why" of UPP substrate ubiquitination and subsequent degradation, and by recycling ubiquitin for re-use by the UPP. Because of the reliance of the UPP on DUB activities, it is not surprising that these proteases play important roles in the normal activities of the nervous system and in neurodegenerative diseases. In this review, we summarize recent advances in understanding the functions of DUBs in the nervous system. We focus on their role in the UPP, and make the argument that understanding the UPP from the perspective of DUBs can yield new insight into diseases that result from anomalous intra-cellular processes or inter-cellular networks. Lastly, we discuss the relevance of DUBs as therapeutic options for disorders of the nervous system.
Keywords: brain; glia; neurodegeneration; neuron; protease; proteasome; synapse; ubiquitin.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

