Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes
- PMID: 25191308
- PMCID: PMC4138442
- DOI: 10.3389/fmicb.2014.00426
Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes
Abstract
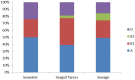
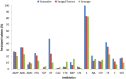
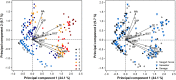
The aim of this study was to examine antibiotic resistance (AR) dissemination in coastal water, considering the contribution of different sources of fecal contamination. Samples were collected in Berlenga, an uninhabited island classified as Natural Reserve and visited by tourists for aquatic recreational activities. To achieve our aim, AR in Escherichia coli isolates from coastal water was compared to AR in isolates from two sources of fecal contamination: human-derived sewage and seagull feces. Isolation of E. coli was done on Chromocult agar. Based on genetic typing 414 strains were established. Distribution of E. coli phylogenetic groups was similar among isolates of all sources. Resistances to streptomycin, tetracycline, cephalothin, and amoxicillin were the most frequent. Higher rates of AR were found among seawater and feces isolates, except for last-line antibiotics used in human medicine. Multi-resistance rates in isolates from sewage and seagull feces (29 and 32%) were lower than in isolates from seawater (39%). Seawater AR profiles were similar to those from seagull feces and differed significantly from sewage AR profiles. Nucleotide sequences matching resistance genes bla TEM, sul1, sul2, tet(A), and tet(B), were present in isolates of all sources. Genes conferring resistance to 3rd generation cephalosporins were detected in seawater (bla CTX-M-1 and bla SHV-12) and seagull feces (bla CMY-2). Plasmid-mediated determinants of resistance to quinolones were found: qnrS1 in all sources and qnrB19 in seawater and seagull feces. Our results show that seawater is a relevant reservoir of AR and that seagulls are an efficient vehicle to spread human-associated bacteria and resistance genes. The E. coli resistome recaptured from Berlenga coastal water was mainly modulated by seagulls-derived fecal pollution. The repertoire of resistance genes covers antibiotics critically important for humans, a potential risk for human health.
Keywords: Escherichia coli; Microbial source tracking; antibiotic resistance; fecal pollution; water quality.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

