Smurf2 E3 ubiquitin ligase modulates proliferation and invasiveness of breast cancer cells in a CNKSR2 dependent manner
- PMID: 25191523
- PMCID: PMC4154384
- DOI: 10.1186/1747-1028-9-2
Smurf2 E3 ubiquitin ligase modulates proliferation and invasiveness of breast cancer cells in a CNKSR2 dependent manner
Abstract
Background: Smurf2 is a member of the HECT family of E3 ubiquitin ligases that play important roles in determining the competence of cells to respond to TGF- β/BMP signaling pathway. However, besides TGF-β/BMP pathway, Smurf2 regulates a repertoire of other signaling pathways ranging from planar cell polarity during embryonic development to cell proliferation, migration, differentiation and senescence. Expression of Smurf2 is found to be dysregulated in many cancers including breast cancer. The purpose of the present study is to examine the effect of Smurf2 knockdown on the tumorigenic potential of human breast cancer cells emphasizing more on proliferative signaling pathway.
Methods: siRNAs targeting different regions of the Smurf2 mRNA were employed to knockdown the expression of Smurf2. The biological effects of synthetic siRNAs on human breast cancer cells were investigated by examining the cell proliferation, migration, invasion, focus formation, anchorage-independent growth, cell cycle arrest, and cell cycle and cell proliferation related protein expressions upon Smurf2 silencing.
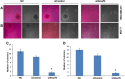
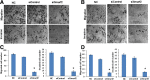
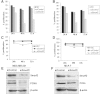
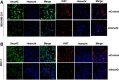
Results: Smurf2 silencing in human breast cancer cells resulted in a decreased focus formation potential and clonogenicity as well as in vitro cell migration/invasion capabilities. Moreover, knockdown of Smurf2 suppressed cell proliferation. Cell cycle analysis showed that the anti-proliferative effect of Smurf2 siRNA was mediated by arresting cells in the G0/G1 phase, which was caused by decreased expression of cyclin D1and cdk4, followed by upregulation p21 and p27. Furthermore, we demonstrated that silencing of Smurf2 downregulated the proliferation of breast cancer cells by modulating the PI3K- PTEN-AKT-FoxO3a pathway via the scaffold protein CNKSR2 which is involved in RAS-dependent signaling pathways. The present study provides the first evidence that silencing Smurf2 using synthetic siRNAs can regulate the tumorigenic properties of human breast cancer cells in a CNKSR2 dependent manner.
Conclusions: Our results therefore suggest a novel relation between Smurf2 and CNKSR2 thereby regulating AKT-dependent cell proliferation and invasion. Owing to the fact that PI3K-AKT signaling is hyperactivated in various human cancers and that Smurf2 also regulates cellular transformation, our results indicate that Smurf2 may serve as a potential molecule for targeted cancer therapy of certain tumour types including breast cancer.
Keywords: Breast cancer; CNKSR2; Oncogenic signaling; PI3K-AKT; Proliferation; Smurf2.
Figures









Similar articles
-
Regulation of CNKSR2 protein stability by the HECT E3 ubiquitin ligase Smurf2, and its role in breast cancer progression.BMC Cancer. 2018 Mar 13;18(1):284. doi: 10.1186/s12885-018-4188-x. BMC Cancer. 2018. PMID: 29534682 Free PMC article.
-
Silencing of the IKKε gene by siRNA inhibits invasiveness and growth of breast cancer cells.Breast Cancer Res. 2010;12(5):R74. doi: 10.1186/bcr2644. Epub 2010 Sep 23. Breast Cancer Res. 2010. PMID: 20863366 Free PMC article.
-
Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells.J Biol Chem. 2008 Dec 19;283(51):35660-7. doi: 10.1074/jbc.M710496200. Epub 2008 Oct 16. J Biol Chem. 2008. PMID: 18927080
-
The functions and regulation of Smurfs in cancers.Semin Cancer Biol. 2020 Dec;67(Pt 2):102-116. doi: 10.1016/j.semcancer.2019.12.023. Epub 2019 Dec 30. Semin Cancer Biol. 2020. PMID: 31899247 Review.
-
Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression.Biochim Biophys Acta. 2013 Jan;1835(1):119-28. doi: 10.1016/j.bbcan.2012.11.003. Epub 2012 Nov 16. Biochim Biophys Acta. 2013. PMID: 23164545 Review.
Cited by
-
Role of Ubiquitination in PTEN Cellular Homeostasis and Its Implications in GB Drug Resistance.Front Oncol. 2020 Sep 2;10:1569. doi: 10.3389/fonc.2020.01569. eCollection 2020. Front Oncol. 2020. PMID: 32984016 Free PMC article. Review.
-
SMG: self-supervised masked graph learning for cancer gene identification.Brief Bioinform. 2023 Sep 22;24(6):bbad406. doi: 10.1093/bib/bbad406. Brief Bioinform. 2023. PMID: 37950905 Free PMC article.
-
Disease-associated synaptic scaffold protein CNK2 modulates PSD size and influences localisation of the regulatory kinase TNIK.Sci Rep. 2020 Mar 31;10(1):5709. doi: 10.1038/s41598-020-62207-4. Sci Rep. 2020. PMID: 32235845 Free PMC article.
-
Functions of CNKSR2 and Its Association with Neurodevelopmental Disorders.Cells. 2022 Jan 17;11(2):303. doi: 10.3390/cells11020303. Cells. 2022. PMID: 35053419 Free PMC article. Review.
-
E3 ubiquitin ligase Smurf2: a prognostic factor in microsatellite stable colorectal cancer.Cancer Manag Res. 2019 Feb 22;11:1795-1803. doi: 10.2147/CMAR.S178111. eCollection 2019. Cancer Manag Res. 2019. PMID: 30863185 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

