The CCA-end of P-tRNA Contacts Both the Human RPL36AL and the A-site Bound Translation Termination Factor eRF1 at the Peptidyl Transferase Center of the Human 80S Ribosome
- PMID: 25191528
- PMCID: PMC4150381
- DOI: 10.2174/1874091X01408010052
The CCA-end of P-tRNA Contacts Both the Human RPL36AL and the A-site Bound Translation Termination Factor eRF1 at the Peptidyl Transferase Center of the Human 80S Ribosome
Abstract
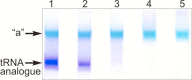
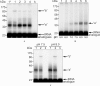
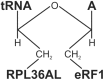
We have demonstrated previously that the E-site specific protein RPL36AL present in human ribosomes can be crosslinked with the CCA-end of a P-tRNA in situ. Here we report the following: (i) We modeled RPL36AL into the structure of the archaeal ortholog RPL44E extracted from the known X-ray structure of the 50S subunit of Haloarcula marismortui. Superimposing the obtained RPL36AL structure with that of P/E tRNA observed in eukaryotic 80S ribosomes suggested that RPL36AL might in addition to its CCA neighbourhood interact with the inner site of the tRNA elbow similar to an interaction pattern known from tRNA•synthetase pairs. (ii) Accordingly, we detected that the isolated recombinant protein RPL36AL can form a tight binary complex with deacylated tRNA, and even tRNA fragments truncated at their CCA end showed a high affinity in the nanomolar range supporting a strong interaction outside the CCA end. (iii) We constructed programmed 80S complexes containing the termination factor eRF1 (stop codon UAA at the A-site) and a 2',3'-dialdehyde tRNA (tRNAox) analog at the P-site. Surprisingly, we observed a crosslinked ternary complex containing the tRNA, eRF1 and RPL36AL crosslinked both to the aldehyde groups of tRNAox at the 2'- and 3'-positions of the ultimate A. We also demonstrated that, upon binding to the ribosomal A-site, eRF1 induces an alternative conformation of the ribosome and/or the tRNA, leading to a novel crosslink of tRNAox to another large-subunit ribosomal protein (namely L37) rather than to RPL36AL, both ribosomal proteins being labeled in a mutually exclusive fashion. Since the human 80S ribosome in complex with P-site bound tRNAox and A-site bound eRF1 corresponds to the post-termination state of the ribosome, the results represent the first biochemical evidence for the positioning of the CCA-arm of the P-tRNA in close proximity to both RPL36AL and eRF1 at the end of the translation process.
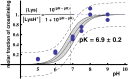
Keywords: A-site stop codon; CCA-end; P-tRNA; RPL36AL/tRNAox/eRF1 ternary complex on the human 80S ribosome.; abnormally low pK for Lys-53 of human RPL36AL; conformational change of eRF1 upon binding to the ribosome; crosslinking; eRF1; effect of the eRF1/eRF3 complex on the crosslinking of eRF1 in the human 80S ribosome; human s80S ribosomes; recombinant human RPL36AL.
Figures







Similar articles
-
The human large subunit ribosomal protein L36A-like contacts the CCA end of P-site bound tRNA.Biochimie. 2009 Nov-Dec;91(11-12):1420-5. doi: 10.1016/j.biochi.2009.07.013. Epub 2009 Jul 31. Biochimie. 2009. PMID: 19647033
-
Exploring contacts of eRF1 with the 3'-terminus of the P site tRNA and mRNA stop signal in the human ribosome at various translation termination steps.Biochim Biophys Acta Gene Regul Mech. 2017 Jul;1860(7):782-793. doi: 10.1016/j.bbagrm.2017.04.004. Epub 2017 Apr 27. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28457996
-
Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes.FEBS Lett. 2002 Mar 6;514(1):96-101. doi: 10.1016/s0014-5793(02)02304-9. FEBS Lett. 2002. PMID: 11904189
-
Termination and post-termination events in eukaryotic translation.Adv Protein Chem Struct Biol. 2012;86:45-93. doi: 10.1016/B978-0-12-386497-0.00002-5. Adv Protein Chem Struct Biol. 2012. PMID: 22243581 Review.
-
Translation Termination and Ribosome Recycling in Eukaryotes.Cold Spring Harb Perspect Biol. 2018 Oct 1;10(10):a032656. doi: 10.1101/cshperspect.a032656. Cold Spring Harb Perspect Biol. 2018. PMID: 29735640 Free PMC article. Review.
Cited by
-
Ribosomal protein paralogues in ribosome specialization.Philos Trans R Soc Lond B Biol Sci. 2025 Mar 6;380(1921):20230387. doi: 10.1098/rstb.2023.0387. Epub 2025 Mar 6. Philos Trans R Soc Lond B Biol Sci. 2025. PMID: 40045786 Review.
-
RpbL12 Assists Catalysis by Correctly Positioning the Incoming Aminoacyl-tRNA in the A-Site of E. coli 70S Ribosomes.Open Biochem J. 2018 Jul 31;12:113-129. doi: 10.2174/1874091X01812010113. eCollection 2018. Open Biochem J. 2018. PMID: 30197688 Free PMC article.
-
Chemical footprinting reveals conformational changes of 18S and 28S rRNAs at different steps of translation termination on the human ribosome.RNA. 2016 Feb;22(2):278-89. doi: 10.1261/rna.053801.115. Epub 2015 Dec 11. RNA. 2016. PMID: 26655225 Free PMC article.
-
Molecular alterations in skeletal muscle in rheumatoid arthritis are related to disease activity, physical inactivity, and disability.Arthritis Res Ther. 2017 Jan 23;19(1):12. doi: 10.1186/s13075-016-1215-7. Arthritis Res Ther. 2017. PMID: 28114971 Free PMC article.
-
A Functional Role for the Monomethylated Gln-51 and Lys-53 Residues of the 49GGQTK53 Motif of eL42 from Human 80S Ribosomes.Open Biochem J. 2017 Mar 31;11:8-26. doi: 10.2174/1874091X01711010008. eCollection 2017. Open Biochem J. 2017. PMID: 28567122 Free PMC article.
References
-
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. - PubMed
-
- Steitz TA, Moore PB. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–418. - PubMed
-
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. - PubMed
-
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. - PubMed
-
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
