Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site
- PMID: 25192068
- PMCID: PMC4156391
- DOI: 10.1371/journal.pone.0107005
Crystal structure of transglutaminase 2 with GTP complex and amino acid sequence evidence of evolution of GTP binding site
Abstract
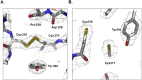
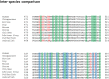
Transglutaminase2 (TG2) is a multi-functional protein involved in various cellular processes, including apoptosis, differentiation, wound healing, and angiogenesis. The malfunction of TG2 causes many human disease including inflammatory disease, celiac disease, neurodegenerative diseases, tissue fibrosis, and cancers. Protein cross-linking activity, which is representative of TG2, is activated by calcium ions and suppressed by GTP. Here, we elucidated the structure of TG2 in complex with its endogenous inhibitor, GTP. Our structure showed why GTP is the optimal nucleotide for interacting with and inhibiting TG2. In addition, sequence comparison provided information describing the evolutionary scenario of GTP usage for controlling the activity of TG2.
Conflict of interest statement
Figures

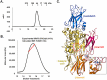



Similar articles
-
Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity.PLoS One. 2018 Oct 15;13(10):e0204707. doi: 10.1371/journal.pone.0204707. eCollection 2018. PLoS One. 2018. PMID: 30321187 Free PMC article.
-
Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2.Int J Mol Sci. 2020 Jan 25;21(3):791. doi: 10.3390/ijms21030791. Int J Mol Sci. 2020. PMID: 31991788 Free PMC article.
-
External GTP-bound transglutaminase 2 is a molecular switch for chondrocyte hypertrophic differentiation and calcification.J Biol Chem. 2005 Apr 15;280(15):15004-12. doi: 10.1074/jbc.M500962200. Epub 2005 Feb 3. J Biol Chem. 2005. PMID: 15691824
-
Structural aspects of transglutaminase 2: functional, structural, and regulatory diversity.Apoptosis. 2017 Sep;22(9):1057-1068. doi: 10.1007/s10495-017-1396-9. Apoptosis. 2017. PMID: 28677093 Review.
-
Transglutaminase 2: an enigmatic enzyme with diverse functions.Trends Biochem Sci. 2002 Oct;27(10):534-9. doi: 10.1016/s0968-0004(02)02182-5. Trends Biochem Sci. 2002. PMID: 12368090 Review.
Cited by
-
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.Commun Biol. 2024 Aug 13;7(1):982. doi: 10.1038/s42003-024-06672-x. Commun Biol. 2024. PMID: 39134806 Free PMC article.
-
Transglutaminase is a tumor cell and cancer stem cell survival factor.Mol Carcinog. 2015 Oct;54(10):947-58. doi: 10.1002/mc.22375. Epub 2015 Aug 10. Mol Carcinog. 2015. PMID: 26258961 Free PMC article. Review.
-
Transamidase site-targeted agents alter the conformation of the transglutaminase cancer stem cell survival protein to reduce GTP binding activity and cancer stem cell survival.Oncogene. 2017 May 25;36(21):2981-2990. doi: 10.1038/onc.2016.452. Epub 2016 Dec 12. Oncogene. 2017. PMID: 27941875 Free PMC article.
-
Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity.PLoS One. 2018 Oct 15;13(10):e0204707. doi: 10.1371/journal.pone.0204707. eCollection 2018. PLoS One. 2018. PMID: 30321187 Free PMC article.
-
Transglutaminase 2-mediated histone monoaminylation and its role in cancer.Biosci Rep. 2024 Aug 28;44(8):BSR20240493. doi: 10.1042/BSR20240493. Biosci Rep. 2024. PMID: 39115570 Free PMC article. Review.
References
-
- Nemes Z Jr, Adany R, Balazs M, Boross P, Fesus L (1997) Identification of cytoplasmic actin as an abundant glutaminyl substrate for tissue transglutaminase in HL-60 and U937 cells undergoing apoptosis. J Biol Chem 272: 20577–20583. - PubMed
-
- Piacentini M, Fesus L, Farrace MG, Ghibelli L, Piredda L, et al. (1991) The expression of “tissue” transglutaminase in two human cancer cell lines is related with the programmed cell death (apoptosis). Eur J Cell Biol 54: 246–254. - PubMed
-
- Jones RA, Kotsakis P, Johnson TS, Chau DY, Ali S, et al. (2006) Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ 13: 1442–1453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

