The developmental potential of iPSCs is greatly influenced by reprogramming factor selection
- PMID: 25192464
- PMCID: PMC4170792
- DOI: 10.1016/j.stem.2014.07.003
The developmental potential of iPSCs is greatly influenced by reprogramming factor selection
Abstract
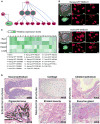
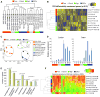
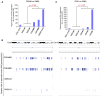
Induced pluripotent stem cells (iPSCs) are commonly generated by transduction of Oct4, Sox2, Klf4, and Myc (OSKM) into cells. Although iPSCs are pluripotent, they frequently exhibit high variation in terms of quality, as measured in mice by chimera contribution and tetraploid complementation. Reliably high-quality iPSCs will be needed for future therapeutic applications. Here, we show that one major determinant of iPSC quality is the combination of reprogramming factors used. Based on tetraploid complementation, we found that ectopic expression of Sall4, Nanog, Esrrb, and Lin28 (SNEL) in mouse embryonic fibroblasts (MEFs) generated high-quality iPSCs more efficiently than other combinations of factors including OSKM. Although differentially methylated regions, transcript number of master regulators, establishment of specific superenhancers, and global aneuploidy were comparable between high- and low-quality lines, aberrant gene expression, trisomy of chromosome 8, and abnormal H2A.X deposition were distinguishing features that could potentially also be applicable to human.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



References
-
- Audo I, Bujakowska KM, Léveillard T, Mohand-Saïd S, Lancelot ME, Germain A, Antonio A, Michiels C, Saraiva JP, Letexier M, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7:8. - PMC - PubMed
-
- Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. - PubMed
-
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. - PubMed
-
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

