The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state
- PMID: 25192989
- PMCID: PMC4249009
- DOI: 10.1128/AEM.01576-14
The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state
Abstract
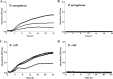
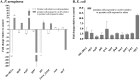
Persister cells, which are tolerant to antimicrobials, contribute to biofilm recalcitrance to therapeutic agents. In turn, the ability to kill persister cells is believed to significantly improve efforts in eradicating biofilm-related, chronic infections. While much research has focused on elucidating the mechanism(s) by which persister cells form, little is known about the mechanism or factors that enable persister cells to revert to an active and susceptible state. Here, we demonstrate that cis-2-decenoic acid (cis-DA), a fatty acid signaling molecule, is able to change the status of Pseudomonas aeruginosa and Escherichia coli persister cells from a dormant to a metabolically active state without an increase in cell number. This cell awakening is supported by an increase of the persister cells' respiratory activity together with changes in protein abundance and increases of the transcript expression levels of several metabolic markers, including acpP, 16S rRNA, atpH, and ppx. Given that most antimicrobials target actively growing cells, we also explored the effect of cis-DA on enhancing antibiotic efficacy in killing persister cells due to their inability to keep a persister cell state. Compared to antimicrobial treatment alone, combinational treatments of persister cell subpopulations with antimicrobials and cis-DA resulted in a significantly greater decrease in cell viability. In addition, the presence of cis-DA led to a decrease in the number of persister cells isolated. We thus demonstrate the ability of a fatty acid signaling molecule to revert bacterial cells from a tolerant phenotype to a metabolically active, antimicrobial-sensitive state.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures








Similar articles
-
Medium chain unsaturated fatty acid ethyl esters inhibit persister formation of Escherichia coli via antitoxin HipB.Appl Microbiol Biotechnol. 2018 Oct;102(19):8511-8524. doi: 10.1007/s00253-018-9271-3. Epub 2018 Aug 7. Appl Microbiol Biotechnol. 2018. PMID: 30088019
-
Interaction of Staphylococcus aureus persister cells with the host when in a persister state and following awakening.Sci Rep. 2016 Aug 10;6:31342. doi: 10.1038/srep31342. Sci Rep. 2016. PMID: 27506163 Free PMC article.
-
Persister cells.Annu Rev Microbiol. 2010;64:357-72. doi: 10.1146/annurev.micro.112408.134306. Annu Rev Microbiol. 2010. PMID: 20528688 Review.
-
Human Granulocyte Macrophage Colony-Stimulating Factor Enhances Antibiotic Susceptibility of Pseudomonas aeruginosa Persister Cells.Sci Rep. 2015 Nov 30;5:17315. doi: 10.1038/srep17315. Sci Rep. 2015. PMID: 26616387 Free PMC article.
-
Control of Biofilms with the Fatty Acid Signaling Molecule cis-2-Decenoic Acid.Pharmaceuticals (Basel). 2015 Nov 25;8(4):816-35. doi: 10.3390/ph8040816. Pharmaceuticals (Basel). 2015. PMID: 26610524 Free PMC article. Review.
Cited by
-
Immune Response Modulation by Pseudomonas aeruginosa Persister Cells.mBio. 2023 Apr 25;14(2):e0005623. doi: 10.1128/mbio.00056-23. Epub 2023 Mar 15. mBio. 2023. PMID: 36920189 Free PMC article.
-
The Open Challenge of in vitro Modeling Complex and Multi-Microbial Communities in Three-Dimensional Niches.Front Bioeng Biotechnol. 2020 Oct 20;8:539319. doi: 10.3389/fbioe.2020.539319. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33195112 Free PMC article. Review.
-
Recent Advances in Bacterial Persistence Mechanisms.Int J Mol Sci. 2023 Sep 20;24(18):14311. doi: 10.3390/ijms241814311. Int J Mol Sci. 2023. PMID: 37762613 Free PMC article. Review.
-
Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis.Front Cell Infect Microbiol. 2022 Jan 6;11:824042. doi: 10.3389/fcimb.2021.824042. eCollection 2021. Front Cell Infect Microbiol. 2022. PMID: 35071057 Free PMC article. Review.
-
Toxin-antitoxin systems and their medical applications: current status and future perspective.Appl Microbiol Biotechnol. 2021 Mar;105(5):1803-1821. doi: 10.1007/s00253-021-11134-z. Epub 2021 Feb 13. Appl Microbiol Biotechnol. 2021. PMID: 33582835 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

