Protein subcellular relocalization of duplicated genes in Arabidopsis
- PMID: 25193306
- PMCID: PMC4202327
- DOI: 10.1093/gbe/evu191
Protein subcellular relocalization of duplicated genes in Arabidopsis
Abstract
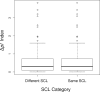
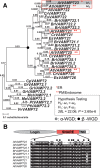
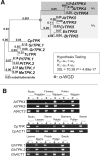
Gene duplications during eukaroytic evolution, by successive rounds of polyploidy and by smaller scale duplications, have provided an enormous reservoir of new genes for the evolution of new functions. Preservation of many duplicated genes can be ascribed to changes in sequences, expression patterns, and functions. Protein subcellular relocalization (protein targeting to a new location within the cell) is another way that duplicated genes can diverge. We studied subcellular relocalization of gene pairs duplicated during the evolution of the Brassicaceae including gene pairs from the alpha whole genome duplication that occurred at the base of the family. We analyzed experimental localization data from green fluorescent protein experiments for 128 duplicate pairs in Arabidopsis thaliana, revealing 19 pairs with subcellular relocalization. Many more of the duplicate pairs with relocalization than with the same localization showed an accelerated rate of amino acid sequence evolution in one duplicate, and one gene showed evidence for positive selection. We studied six duplicate gene pairs in more detail. We used gene family analysis with several pairs to infer which gene shows relocalization. We identified potential sequence mutations through comparative analysis that likely result in relocalization of two duplicated gene products. We show that four cases of relocalization have new expression patterns, compared with orthologs in outgroup species, including two with novel expression in pollen. This study provides insights into subcellular relocalization of evolutionarily recent gene duplicates and features of genes whose products have been relocalized.
Keywords: gene duplication; subcellular localization; whole genome duplication.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





Similar articles
-
Dramatic change in function and expression pattern of a gene duplicated by polyploidy created a paternal effect gene in the Brassicaceae.Mol Biol Evol. 2010 Dec;27(12):2817-28. doi: 10.1093/molbev/msq169. Epub 2010 Jul 8. Mol Biol Evol. 2010. PMID: 20616146
-
Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis.Mol Biol Evol. 2010 Jul;27(7):1686-97. doi: 10.1093/molbev/msq054. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185454
-
Frequent changes in expression profile and accelerated sequence evolution of duplicated imprinted genes in arabidopsis.Genome Biol Evol. 2014 Jul;6(7):1830-42. doi: 10.1093/gbe/evu144. Genome Biol Evol. 2014. PMID: 25115008 Free PMC article.
-
Gene duplication within the Green Lineage: the case of TEL genes.J Exp Bot. 2012 Sep;63(14):5061-77. doi: 10.1093/jxb/ers181. Epub 2012 Aug 3. J Exp Bot. 2012. PMID: 22865910 Review.
-
Lost in translation: What we have learned from attributes that do not translate from Arabidopsis to other plants.Plant Cell. 2025 May 9;37(5):koaf036. doi: 10.1093/plcell/koaf036. Plant Cell. 2025. PMID: 40371945 Free PMC article. Review.
Cited by
-
Comprehensive analysis of Lon proteases in plants highlights independent gene duplication events.J Exp Bot. 2019 Apr 12;70(7):2185-2197. doi: 10.1093/jxb/ery440. J Exp Bot. 2019. PMID: 30590727 Free PMC article.
-
Atypical Protein Phosphatase 2A Gene Families Do Not Expand via Paleopolyploidization.Plant Physiol. 2017 Feb;173(2):1283-1300. doi: 10.1104/pp.16.01768. Epub 2016 Dec 29. Plant Physiol. 2017. PMID: 28034953 Free PMC article.
-
Genome-Wide Comparative Analysis of R2R3 MYB Gene Family in Populus and Salix and Identification of Male Flower Bud Development-Related Genes.Front Plant Sci. 2021 Sep 14;12:721558. doi: 10.3389/fpls.2021.721558. eCollection 2021. Front Plant Sci. 2021. PMID: 34594352 Free PMC article.
-
Gene Duplication Accelerates the Pace of Protein Gain and Loss from Plant Organelles.Mol Biol Evol. 2020 Apr 1;37(4):969-981. doi: 10.1093/molbev/msz275. Mol Biol Evol. 2020. PMID: 31750917 Free PMC article.
-
A metabolic, phylogenomic and environmental atlas of diatom plastid transporters from the model species Phaeodactylum.Front Plant Sci. 2022 Sep 22;13:950467. doi: 10.3389/fpls.2022.950467. eCollection 2022. Front Plant Sci. 2022. PMID: 36212359 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

