De novo transcriptome sequence assembly from coconut leaves and seeds with a focus on factors involved in RNA-directed DNA methylation
- PMID: 25193496
- PMCID: PMC4232540
- DOI: 10.1534/g3.114.013409
De novo transcriptome sequence assembly from coconut leaves and seeds with a focus on factors involved in RNA-directed DNA methylation
Abstract
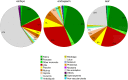
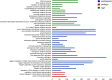
Coconut palm (Cocos nucifera) is a symbol of the tropics and a source of numerous edible and nonedible products of economic value. Despite its nutritional and industrial significance, coconut remains under-represented in public repositories for genomic and transcriptomic data. We report de novo transcript assembly from RNA-seq data and analysis of gene expression in seed tissues (embryo and endosperm) and leaves of a dwarf coconut variety. Assembly of 10 GB sequencing data for each tissue resulted in 58,211 total unigenes in embryo, 61,152 in endosperm, and 33,446 in leaf. Within each unigene pool, 24,857 could be annotated in embryo, 29,731 could be annotated in endosperm, and 26,064 could be annotated in leaf. A KEGG analysis identified 138, 138, and 139 pathways, respectively, in transcriptomes of embryo, endosperm, and leaf tissues. Given the extraordinarily large size of coconut seeds and the importance of small RNA-mediated epigenetic regulation during seed development in model plants, we used homology searches to identify putative homologs of factors required for RNA-directed DNA methylation in coconut. The findings suggest that RNA-directed DNA methylation is important during coconut seed development, particularly in maturing endosperm. This dataset will expand the genomics resources available for coconut and provide a foundation for more detailed analyses that may assist molecular breeding strategies aimed at improving this major tropical crop.
Keywords: RNA-seq; coconut; endosperm; epigenetics; monocot.
Copyright © 2014 Huang et al.
Figures




References
-
- Abraham A., Mathew P. M., 1963. Cytology of coconut endosperm. Ann. Bot. (Lond.) 27: 505–512.
-
- Al-Dous E. K., George B., Al-Mahmoud M. E., Al-Jaber M. Y., Wang H., et al. , 2011. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 29: 521–527. - PubMed
-
- Amson R., Pece S., Marine J. C., Di Fiore P. P., Telerman A., 2013. TPT1/ TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol. 23: 37–46. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
