Ring finger protein 34 (RNF34) interacts with and promotes γ-aminobutyric acid type-A receptor degradation via ubiquitination of the γ2 subunit
- PMID: 25193658
- PMCID: PMC4200290
- DOI: 10.1074/jbc.M114.603068
Ring finger protein 34 (RNF34) interacts with and promotes γ-aminobutyric acid type-A receptor degradation via ubiquitination of the γ2 subunit
Abstract
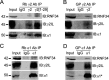
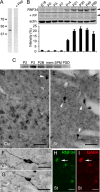
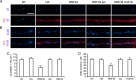
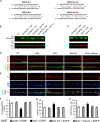
We have found that the large intracellular loop of the γ2 GABAA receptor (R) subunit (γ2IL) interacts with RNF34 (an E3 ubiquitin ligase), as shown by yeast two-hybrid and in vitro pulldown assays. In brain extracts, RNF34 co-immunoprecipitates with assembled GABAARs. In co-transfected HEK293 cells, RNF34 reduces the expression of the γ2 GABAAR subunit by increasing the ratio of ubiquitinated/nonubiquitinated γ2. Mutating several lysines of the γ2IL into arginines makes the γ2 subunit resistant to RNF34-induced degradation. RNF34 also reduces the expression of the γ2 subunit when α1 and β3 subunits are co-assembled with γ2. This effect is partially reversed by leupeptin or MG132, indicating that both the lysosomal and proteasomal degradation pathways are involved. Immunofluorescence of cultured hippocampal neurons shows that RNF34 forms clusters and that a subset of these clusters is associated with GABAergic synapses. This association is also observed in the intact rat brain by electron microscopy immunocytochemistry. RNF34 is not expressed until the 2nd postnatal week of rat brain development, being highly expressed in some interneurons. Overexpression of RNF34 in hippocampal neurons decreases the density of γ2 GABAAR clusters and the number of GABAergic contacts that these neurons receive. Knocking down endogenous RNF34 with shRNA leads to increased γ2 GABAAR cluster density and GABAergic innervation. The results indicate that RNF34 regulates postsynaptic γ2-GABAAR clustering and GABAergic synaptic innervation by interacting with and ubiquitinating the γ2-GABAAR subunit promoting GABAAR degradation.
Keywords: E3 Ubiquitin Ligase; GABAA Receptors; Interaction; Lysosome; Proteasome; RNF34; Synapse; Ubiquitination; Yeast Two-hybrid.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





Similar articles
-
Molecular and functional interaction between protocadherin-γC5 and GABAA receptors.J Neurosci. 2012 Aug 22;32(34):11780-97. doi: 10.1523/JNEUROSCI.0969-12.2012. J Neurosci. 2012. PMID: 22915120 Free PMC article.
-
Disruption of postsynaptic GABA receptor clusters leads to decreased GABAergic innervation of pyramidal neurons.J Neurochem. 2005 Nov;95(3):756-70. doi: 10.1111/j.1471-4159.2005.03426.x. J Neurochem. 2005. PMID: 16248887
-
RNF34 is a cold-regulated E3 ubiquitin ligase for PGC-1α and modulates brown fat cell metabolism.Mol Cell Biol. 2012 Jan;32(2):266-75. doi: 10.1128/MCB.05674-11. Epub 2011 Nov 7. Mol Cell Biol. 2012. PMID: 22064484 Free PMC article.
-
Proteostasis regulation of GABAA receptors in neuronal function and disease.Biomed Pharmacother. 2025 May;186:117992. doi: 10.1016/j.biopha.2025.117992. Epub 2025 Mar 20. Biomed Pharmacother. 2025. PMID: 40112516 Free PMC article. Review.
-
Roles of ubiquitination at the synapse.Biochim Biophys Acta. 2008 Aug;1779(8):495-506. doi: 10.1016/j.bbagrm.2007.12.010. Epub 2008 Jan 5. Biochim Biophys Acta. 2008. PMID: 18222124 Free PMC article. Review.
Cited by
-
Grp94 Protein Delivers γ-Aminobutyric Acid Type A (GABAA) Receptors to Hrd1 Protein-mediated Endoplasmic Reticulum-associated Degradation.J Biol Chem. 2016 Apr 29;291(18):9526-39. doi: 10.1074/jbc.M115.705004. Epub 2016 Mar 4. J Biol Chem. 2016. PMID: 26945068 Free PMC article.
-
In vivo transgenic expression of collybistin in neurons of the rat cerebral cortex.J Comp Neurol. 2017 Apr 1;525(5):1291-1311. doi: 10.1002/cne.24137. Epub 2016 Nov 21. J Comp Neurol. 2017. PMID: 27804142 Free PMC article.
-
Modulating Endoplasmic Reticulum Chaperones and Mutant Protein Degradation in GABRG2(Q390X) Associated with Genetic Epilepsy with Febrile Seizures Plus and Dravet Syndrome.Int J Mol Sci. 2024 Apr 23;25(9):4601. doi: 10.3390/ijms25094601. Int J Mol Sci. 2024. PMID: 38731820 Free PMC article.
-
Expression of protocadherin-γC4 protein in the rat brain.J Comp Neurol. 2020 Apr 1;528(5):840-864. doi: 10.1002/cne.24783. Epub 2019 Nov 6. J Comp Neurol. 2020. PMID: 31609469 Free PMC article.
-
Influence of APOE and RNF219 on Behavioral and Cognitive Features of Female Patients Affected by Mild Cognitive Impairment or Alzheimer's Disease.Front Aging Neurosci. 2018 Apr 13;10:92. doi: 10.3389/fnagi.2018.00092. eCollection 2018. Front Aging Neurosci. 2018. PMID: 29755337 Free PMC article.
References
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Hoppe T. (2005) Multiubiquitylation by E4 enzymes: “one size” doesn't fit all. Trends Biochem. Sci. 30, 183–187 - PubMed
-
- Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 - PubMed
-
- Deshaies R. J., Joazeiro C. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

