Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity
- PMID: 25194926
- PMCID: PMC4183562
- DOI: 10.1038/ncb3028
Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity
Abstract
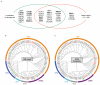
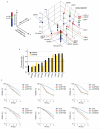
DNA double-strand breaks (DSBs) are perhaps the most toxic of all DNA lesions, with defects in the DNA-damage response to DSBs being associated with various human diseases. Although it is known that DSB repair pathways are tightly regulated by ubiquitylation, we do not yet have a comprehensive understanding of how deubiquitylating enzymes (DUBs) function in DSB responses. Here, by carrying out a multidimensional screening strategy for human DUBs, we identify several with hitherto unknown links to DSB repair, the G2/M DNA-damage checkpoint and genome-integrity maintenance. Phylogenetic analyses reveal functional clustering within certain DUB subgroups, suggesting evolutionally conserved functions and/or related modes of action. Furthermore, we establish that the DUB UCHL5 regulates DSB resection and repair by homologous recombination through protecting its interactor, NFRKB, from degradation. Collectively, our findings extend the list of DUBs promoting the maintenance of genome integrity, and highlight their potential as therapeutic targets for cancer.
Figures





References
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. - PubMed
-
- Kastan MB. DNA damage responses: mechanisms and roles in human disease: 2007 G.H.A. Clowes Memorial Award Lecture. Mol Cancer Res. 2008;6:517–524. - PubMed
-
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. - PubMed
-
- Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

