nDNA-Prot: identification of DNA-binding proteins based on unbalanced classification
- PMID: 25196432
- PMCID: PMC4165999
- DOI: 10.1186/1471-2105-15-298
nDNA-Prot: identification of DNA-binding proteins based on unbalanced classification
Abstract
Background: DNA-binding proteins are vital for the study of cellular processes. In recent genome engineering studies, the identification of proteins with certain functions has become increasingly important and needs to be performed rapidly and efficiently. In previous years, several approaches have been developed to improve the identification of DNA-binding proteins. However, the currently available resources are insufficient to accurately identify these proteins. Because of this, the previous research has been limited by the relatively unbalanced accuracy rate and the low identification success of the current methods.
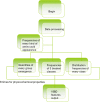
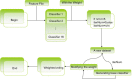
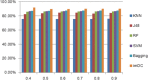
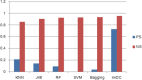
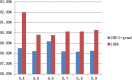
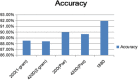
Results: In this paper, we explored the practicality of modelling DNA binding identification and simultaneously employed an ensemble classifier, and a new predictor (nDNA-Prot) was designed. The presented framework is comprised of two stages: a 188-dimension feature extraction method to obtain the protein structure and an ensemble classifier designated as imDC. Experiments using different datasets showed that our method is more successful than the traditional methods in identifying DNA-binding proteins. The identification was conducted using a feature that selected the minimum Redundancy and Maximum Relevance (mRMR). An accuracy rate of 95.80% and an Area Under the Curve (AUC) value of 0.986 were obtained in a cross validation. A test dataset was tested in our method and resulted in an 86% accuracy, versus a 76% using iDNA-Prot and a 68% accuracy using DNA-Prot.
Conclusions: Our method can help to accurately identify DNA-binding proteins, and the web server is accessible at http://datamining.xmu.edu.cn/~songli/nDNA. In addition, we also predicted possible DNA-binding protein sequences in all of the sequences from the UniProtKB/Swiss-Prot database.
Figures


References
-
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. Uniprotkb/swiss-prot. Plant Bioinformatics. Humana Press. 2007;406:89–112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

