Multiscale digital Arabidopsis predicts individual organ and whole-organism growth
- PMID: 25197087
- PMCID: PMC4191812
- DOI: 10.1073/pnas.1410238111
Multiscale digital Arabidopsis predicts individual organ and whole-organism growth
Erratum in
-
Correction for Chew et al., Multiscale digital Arabidopsis predicts individual organ and whole-organism growth.Proc Natl Acad Sci U S A. 2015 May 12;112(19):E2556. doi: 10.1073/pnas.1506983112. Epub 2015 Apr 22. Proc Natl Acad Sci U S A. 2015. PMID: 25902547 Free PMC article. No abstract available.
Abstract
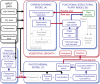
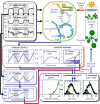
Understanding how dynamic molecular networks affect whole-organism physiology, analogous to mapping genotype to phenotype, remains a key challenge in biology. Quantitative models that represent processes at multiple scales and link understanding from several research domains can help to tackle this problem. Such integrated models are more common in crop science and ecophysiology than in the research communities that elucidate molecular networks. Several laboratories have modeled particular aspects of growth in Arabidopsis thaliana, but it was unclear whether these existing models could productively be combined. We test this approach by constructing a multiscale model of Arabidopsis rosette growth. Four existing models were integrated with minimal parameter modification (leaf water content and one flowering parameter used measured data). The resulting framework model links genetic regulation and biochemical dynamics to events at the organ and whole-plant levels, helping to understand the combined effects of endogenous and environmental regulators on Arabidopsis growth. The framework model was validated and tested with metabolic, physiological, and biomass data from two laboratories, for five photoperiods, three accessions, and a transgenic line, highlighting the plasticity of plant growth strategies. The model was extended to include stochastic development. Model simulations gave insight into the developmental control of leaf production and provided a quantitative explanation for the pleiotropic developmental phenotype caused by overexpression of miR156, which was an open question. Modular, multiscale models, assembling knowledge from systems biology to ecophysiology, will help to understand and to engineer plant behavior from the genome to the field.
Keywords: crop modeling; digital organism; ecology; plant growth model.
Conflict of interest statement
Conflict of interest statement: J.T. and R.M. are directors, shareholders, and employees of Simulistics Ltd. and developers of the Simile software application. Simile was used to visualize a version of the model described. The reference version is in
Figures



References
-
- Chew YH, et al. An augmented Arabidopsis phenology model reveals seasonal temperature control of flowering time. New Phytol. 2012;194(3):654–665. - PubMed
-
- Franklin KA. Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol. 2009;12(1):63–68. - PubMed
-
- Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

