Pharmacogenomic approach to identify drug sensitivity in small-cell lung cancer
- PMID: 25198282
- PMCID: PMC4157793
- DOI: 10.1371/journal.pone.0106784
Pharmacogenomic approach to identify drug sensitivity in small-cell lung cancer
Abstract
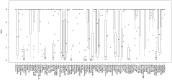
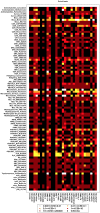
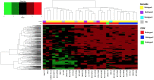
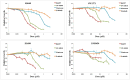
There are currently no molecular targeted approaches to treat small-cell lung cancer (SCLC) similar to those used successfully against non-small-cell lung cancer. This failure is attributable to our inability to identify clinically-relevant subtypes of this disease. Thus, a more systematic approach to drug discovery for SCLC is needed. In this regard, two comprehensive studies recently published in Nature, the Cancer Cell Line Encyclopedia and the Cancer Genome Project, provide a wealth of data regarding the drug sensitivity and genomic profiles of many different types of cancer cells. In the present study we have mined these two studies for new therapeutic agents for SCLC and identified heat shock proteins, cyclin-dependent kinases and polo-like kinases (PLK) as attractive molecular targets with little current clinical trial activity in SCLC. Remarkably, our analyses demonstrated that most SCLC cell lines clustered into a single, predominant subgroup by either gene expression or CNV analyses, leading us to take a pharmacogenomic approach to identify subgroups of drug-sensitive SCLC cells. Using PLK inhibitors as an example, we identified and validated a gene signature for drug sensitivity in SCLC cell lines. This gene signature could distinguish subpopulations among human SCLC tumors, suggesting its potential clinical utility. Finally, circos plots were constructed to yield a comprehensive view of how transcriptional, copy number and mutational elements affect PLK sensitivity in SCLC cell lines. Taken together, this study outlines an approach to predict drug sensitivity in SCLC to novel targeted therapeutics.
Conflict of interest statement
Figures


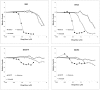
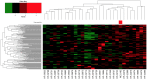

References
-
- Jackman DM, Johnson BE (2005) Small-cell lung cancer. The Lancet 366: 1385–1396. - PubMed
-
- D'Angelo SP, Pietanza MC (2010) The molecular pathogenesis of small cell lung cancer. Cancer Biology & Therapy 10: 1–10. - PubMed
-
- Hurwitz JL, McCoy F, Scullin P, Fennell DA (2009) New advances in the second-line treatment of small cell lung cancer. The Oncologist 14: 986–994. - PubMed
-
- William WN, Glisson BS (2011) Novel strategies for the treatment of small-cell lung carcinoma. Nat Rev Clin Oncol 8: 611–619. - PubMed
-
- Nickolich M, Babakoohi S, Fu P, Dowlati A (2014) Clinical Trial Design in Small Cell Lung Cancer: Surrogate End Points and Statistical Evolution. Clin Lung Cancer 15: 207–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

