Cellular, molecular and functional characterisation of YAC transgenic mouse models of Friedreich ataxia
- PMID: 25198290
- PMCID: PMC4157886
- DOI: 10.1371/journal.pone.0107416
Cellular, molecular and functional characterisation of YAC transgenic mouse models of Friedreich ataxia
Abstract
Background: Friedreich ataxia (FRDA) is an autosomal recessive neurodegenerative disorder, caused by a GAA repeat expansion mutation within intron 1 of the FXN gene. We have previously established and performed preliminary characterisation of several human FXN yeast artificial chromosome (YAC) transgenic FRDA mouse models containing GAA repeat expansions, Y47R (9 GAA repeats), YG8R (90 and 190 GAA repeats) and YG22R (190 GAA repeats).
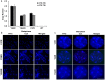
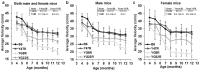
Methodology/principal findings: We now report extended cellular, molecular and functional characterisation of these FXN YAC transgenic mouse models. FXN transgene copy number analysis of the FRDA mice demonstrated that the YG22R and Y47R lines each have a single copy of the FXN transgene while the YG8R line has two copies. Single integration sites of all transgenes were confirmed by fluorescence in situ hybridisation (FISH) analysis of metaphase and interphase chromosomes. We identified significant functional deficits, together with a degree of glucose intolerance and insulin hypersensitivity, in YG8R and YG22R FRDA mice compared to Y47R and wild-type control mice. We also confirmed increased somatic GAA repeat instability in the cerebellum and brain of YG22R and YG8R mice, together with significantly reduced levels of FXN mRNA and protein in the brain and liver of YG8R and YG22R compared to Y47R.
Conclusions/significance: Together these studies provide a detailed characterisation of our GAA repeat expansion-based YAC transgenic FRDA mouse models that will help investigations of FRDA disease mechanisms and therapy.
Conflict of interest statement
Figures







References
-
- Pandolfo M (2002) The molecular basis of Friedreich ataxia. Adv Exp Med Biol 516: 99–118. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

