PTEN interacts with histone H1 and controls chromatin condensation
- PMID: 25199838
- PMCID: PMC4201947
- DOI: 10.1016/j.celrep.2014.08.008
PTEN interacts with histone H1 and controls chromatin condensation
Abstract
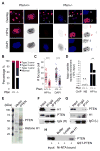
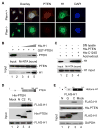
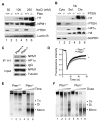
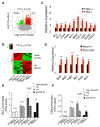
Chromatin organization and dynamics are integral to global gene transcription. Histone modification influences chromatin status and gene expression. PTEN plays multiple roles in tumor suppression, development, and metabolism. Here, we report on the interplay of PTEN, histone H1, and chromatin. We show that loss of PTEN leads to dissociation of histone H1 from chromatin and decondensation of chromatin. PTEN deletion also results in elevation of histone H4 acetylation at lysine 16, an epigenetic marker for chromatin activation. We found that PTEN and histone H1 physically interact through their C-terminal domains. Disruption of the PTEN C terminus promotes the chromatin association of MOF acetyltransferase and induces H4K16 acetylation. Hyperacetylation of H4K16 impairs the association of PTEN with histone H1, which constitutes regulatory feedback that may reduce chromatin stability. Our results demonstrate that PTEN controls chromatin condensation, thus influencing gene expression. We propose that PTEN regulates global gene transcription profiling through histones and chromatin remodeling.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
Chromatin: Interplay of PTEN with histone H1.Nat Rev Mol Cell Biol. 2014 Oct;15(10):630. doi: 10.1038/nrm3883. Epub 2014 Sep 17. Nat Rev Mol Cell Biol. 2014. PMID: 25237823 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

