Synonymous codon bias and functional constraint on GC3-related DNA backbone dynamics in the prokaryotic nucleoid
- PMID: 25200075
- PMCID: PMC4176184
- DOI: 10.1093/nar/gku811
Synonymous codon bias and functional constraint on GC3-related DNA backbone dynamics in the prokaryotic nucleoid
Abstract
While mRNA stability has been demonstrated to control rates of translation, generating both global and local synonymous codon biases in many unicellular organisms, this explanation cannot adequately explain why codon bias strongly tracks neighboring intergene GC content; suggesting that structural dynamics of DNA might also influence codon choice. Because minor groove width is highly governed by 3-base periodicity in GC, the existence of triplet-based codons might imply a functional role for the optimization of local DNA molecular dynamics via GC content at synonymous sites (≈GC3). We confirm a strong association between GC3-related intrinsic DNA flexibility and codon bias across 24 different prokaryotic multiple whole-genome alignments. We develop a novel test of natural selection targeting synonymous sites and demonstrate that GC3-related DNA backbone dynamics have been subject to moderate selective pressure, perhaps contributing to our observation that many genes possess extreme DNA backbone dynamics for their given protein space. This dual function of codons may impose universal functional constraints affecting the evolution of synonymous and non-synonymous sites. We propose that synonymous sites may have evolved as an 'accessory' during an early expansion of a primordial genetic code, allowing for multiplexed protein coding and structural dynamic information within the same molecular context.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

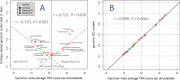


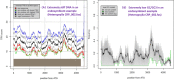
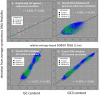

References
-
- Roth A.C., Anisimova M., Cannarrozzi G. Measuring codon usage bias. In: Cannarrozzi G, Schneider A, editors. Codon Evolution: Mechanisms and Models. Oxford: Oxford University Press; 2012. pp. 189–217.
-
- Rubenstein N.D., Pupko T. Detection and analysis of conservation at synonymous sites. In: Cannarrozzi G, Schneider A., editors. Codon Evolution: Mechanisms and Models. Oxford: Oxford University Press; 2012. pp. 218–228.
-
- Zuckerkandl E., Pauling L. Molecules as documents of evolutionary history. J. Theor. Biol. 1965;8:357–366. - PubMed
-
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

