Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation
- PMID: 25200080
- PMCID: PMC4176160
- DOI: 10.1093/nar/gku752
Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation
Abstract
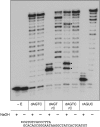
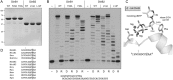
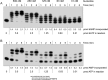
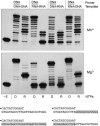
This study unveils Mycobacterium smegmatis DinB2 as the founder of a clade of Y-family DNA polymerase that is naturally adept at incorporating ribonucleotides by virtue of a leucine in lieu of a canonical aromatic steric gate. DinB2 efficiently scavenges limiting dNTP and rNTP substrates in the presence of manganese. DinB2's sugar selectivity factor, gauged by rates of manganese-dependent dNMP versus rNMP addition, is 2.7- to 3.8-fold. DinB2 embeds ribonucleotides during DNA synthesis when rCTP and dCTP are at equimolar concentration. DinB2 can incorporate at least 16 consecutive ribonucleotides. In magnesium, DinB2 has a 26- to 78-fold lower affinity for rNTPs than dNTPs, but only a 2.6- to 6-fold differential in rates of deoxy versus ribo addition (kpol). Two other M. smegmatis Y-family polymerases, DinB1 and DinB3, are characterized here as template-dependent DNA polymerases that discriminate strongly against ribonucleotides, a property that, in the case of DinB1, correlates with its aromatic steric gate side chain. We speculate that the unique ability of DinB2 to utilize rNTPs might allow for DNA repair with a 'ribo patch' when dNTPs are limiting. Phylogenetic analysis reveals DinB2-like polymerases, with leucine, isoleucine or valine steric gates, in many taxa of the phylum Actinobacteria.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Gong C., Martins A., Bongiorno P., Glickman M., Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. - PubMed
-
- Gong C., Bongiorno P., Martins A., Stephanou N.C., Zhu H., Shuman S., Glickman M. S. Mechanism of non-homologous end joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005;12:304–312. - PubMed
-
- Akey D., Martins A., Aniukwu J., Glickman M.S., Shuman S., Berger J.M. Crystal structure and nonhomologous end joining function of the ligase domain of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. - PubMed
-
- Sinha K.M., Stephanou N.C., Gao F., Glickman M.S., Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 2007;282:15114–15125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

