Multifaceted role of the Topo IIIα-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection
- PMID: 25200081
- PMCID: PMC4176181
- DOI: 10.1093/nar/gku803
Multifaceted role of the Topo IIIα-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection
Abstract
BLM, a RecQ family DNA helicase mutated in Bloom's Syndrome, participates in homologous recombination at two stages: 5' DNA end resection and double Holliday junction dissolution. BLM exists in a complex with Topo IIIα, RMI1 and RMI2. Herein, we address the role of Topo IIIα and RMI1-RMI2 in resection using a reconstituted system with purified human proteins. We show that Topo IIIα stimulates DNA unwinding by BLM in a manner that is potentiated by RMI1-RMI2, and that the processivity of resection is reliant on the Topo IIIα-RMI1-RMI2 complex. Topo IIIα localizes to the ends of double-strand breaks, thus implicating it in the recruitment of resection factors. While the single-stranded DNA binding protein RPA plays a major role in imposing the 5' to 3' polarity of resection, Topo IIIα also makes a contribution in this regard. Moreover, we show that DNA2 stimulates the helicase activity of BLM. Our results thus uncover a multifaceted role of the Topo IIIα-RMI1-RMI2 ensemble and of DNA2 in the DNA resection reaction.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

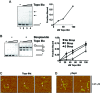

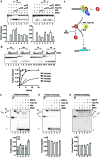
Similar articles
-
Role of replication protein A in double holliday junction dissolution mediated by the BLM-Topo IIIα-RMI1-RMI2 protein complex.J Biol Chem. 2013 May 17;288(20):14221-14227. doi: 10.1074/jbc.M113.465609. Epub 2013 Mar 30. J Biol Chem. 2013. PMID: 23543748 Free PMC article.
-
BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome.Genes Dev. 2008 Oct 15;22(20):2856-68. doi: 10.1101/gad.1725108. Genes Dev. 2008. PMID: 18923083 Free PMC article.
-
Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex.J Biol Chem. 2007 Oct 26;282(43):31484-92. doi: 10.1074/jbc.M706116200. Epub 2007 Aug 28. J Biol Chem. 2007. PMID: 17728255
-
Resolution of Recombination Intermediates: Mechanisms and Regulation.Cold Spring Harb Symp Quant Biol. 2015;80:103-9. doi: 10.1101/sqb.2015.80.027649. Epub 2015 Sep 14. Cold Spring Harb Symp Quant Biol. 2015. PMID: 26370409 Review.
-
The dissolution of double Holliday junctions.Cold Spring Harb Perspect Biol. 2014 Jul 1;6(7):a016477. doi: 10.1101/cshperspect.a016477. Cold Spring Harb Perspect Biol. 2014. PMID: 24984776 Free PMC article. Review.
Cited by
-
Transcription/Replication Conflicts in Tumorigenesis and Their Potential Role as Novel Therapeutic Targets in Multiple Myeloma.Cancers (Basel). 2021 Jul 27;13(15):3755. doi: 10.3390/cancers13153755. Cancers (Basel). 2021. PMID: 34359660 Free PMC article. Review.
-
Accumulation and Phosphorylation of RecQ-Mediated Genome Instability Protein 1 (RMI1) at Serine 284 and Serine 292 during Mitosis.Int J Mol Sci. 2015 Nov 4;16(11):26395-405. doi: 10.3390/ijms161125965. Int J Mol Sci. 2015. PMID: 26556339 Free PMC article.
-
Inhibition of DNA2 nuclease as a therapeutic strategy targeting replication stress in cancer cells.Oncogenesis. 2017 Apr 17;6(4):e319. doi: 10.1038/oncsis.2017.15. Oncogenesis. 2017. PMID: 28414320 Free PMC article.
-
Identification and external validation of a prognostic signature associated with DNA repair genes in gastric cancer.Sci Rep. 2021 Mar 30;11(1):7141. doi: 10.1038/s41598-021-86504-8. Sci Rep. 2021. PMID: 33785812 Free PMC article.
-
Single-cell transcription profiles in Bloom syndrome patients link BLM deficiency with altered condensin complex expression signatures.Hum Mol Genet. 2022 Jul 7;31(13):2185-2193. doi: 10.1093/hmg/ddab373. Hum Mol Genet. 2022. PMID: 35099000 Free PMC article.
References
-
- Daley J.M., Laan R.L., Suresh A., Wilson T.E. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J. Biol. Chem. 2005;280:29030–29037. - PubMed
-
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

